Publications
The Montelione Lab is committed to advancing the frontiers of chemistry. Below is a curated list of our publications, reflecting our lab members’ ongoing contributions and research efforts.
2025
- Gutsche I, Montelione GT. Editorial Overview: Biophysical Methods: Multiple structures of proteins underpin their biological functions. Curr. Opin. Struct. Biol. 84. 102762. 2024.
- Klukowski P, Damberger FF, Allain FHT, Iwai H, Kadavath H, Ramelot TA, Montelione GT, Riek R, Güntert P. The 100-protein nmr spectra dataset: A resource for biomolecular nmr data analysis. Scientific Data. 2024, 11:30.
- Baskaran H, Ploskon E, Tejero R, Yokochi M, Harrus D, Liang Y, Peisach E, Persikova I, Ramelot TA , Sekharan M, …Montelione GT*, Vuister GW*, Young JY*. Restraint validation of biomolecular structures determined by NMR in the Protein Data Bank. Structure (Cell Press) 2024, 32, 824-837. e1. https://doi.org/10.1016/j.str.2024.02.011.
- Mazzei L, Ranieri S, Greene-Cramer R, Cioffi C, Montelione GT, Ciurli S. An isothermal calorimetry assay for determining steady state kinetic and Ensitrelvir inhibition parameters for SARS-CoV-2 3CL-protease. Scientific Reports (Nature Press) 2024, 14, 32175. https://doi.org/10.1038/s41598-024-81990-y
- Mondal A, Singh B, Felkner RH, De Falco A, Swapna G, Montelione GT, Roth MJ, Perez A (2024) A computational pipeline for accurate prioritization of protein-protein binding candidates in high-throughput protein libraries. Angew Chem Int Ed Engl 2024, 63:e202405767. https://doi.org/10.1002/anie.202405767.
- Moyer AP, Ramelot TA, Curti M, Eastman MA, Kang A, Bera AK, Tejero R, Salveson PJ, Curutchet C, Romero E, Montelione GT, Baker D. Enumerative discovery of noncanonical polypeptide secondary structures. J Am Chem Soc 2024, 146: 25501-25512. https://pubs.acs.org/doi/full/10.1021/jacs.4c04991.
- Crouch CC, Chatterjee AK, McCarty C, Song L, Chu A, Johnson K, Heacock M, Riva L, McNamara CL, Wolff KC, Greene-Cramer R, De Falco A, Montelione GT, Grabovyi GA. Synthesis and structure-activity relationship of covalent inhibitors of SARS-CoV-2 papain-like protease with antiviral potency, Bioorg & Med Chem Letts 2025, 16, 130034. https://doi.org/10.1016/j.bmcl.2024.130034
- Elofsson A, Kretsch RC, Magnus M, Montelione GT 2025 Engaging the community: CASP special interest groups. Proteins. 2025, https://doi.org/10.1002/prot.26833
- De Falco A, Greene-Cramer R, Shurina BA, Zakian S, Acton TB, Ramelot TA, Montelione GT. Protocol for production and characterization of SARS-CoV-2 PLpro in Escherichia coli. STAR Protocols 2025, 6, 103952. https://doi.org/10.1016/j.xpro.2025.103955.
- Wu K, Jiang H, Hicks DR, Liu C, Muratspahić E, Ramelot TA, Liu Y, …Gaur A, …Gelb MH, Montelione GT, Derivery E, Baker D. Design of intrinsically disordered region binding proteins. Science 2025, 389, https://doi.org/10.1126/science.adr80.
2024
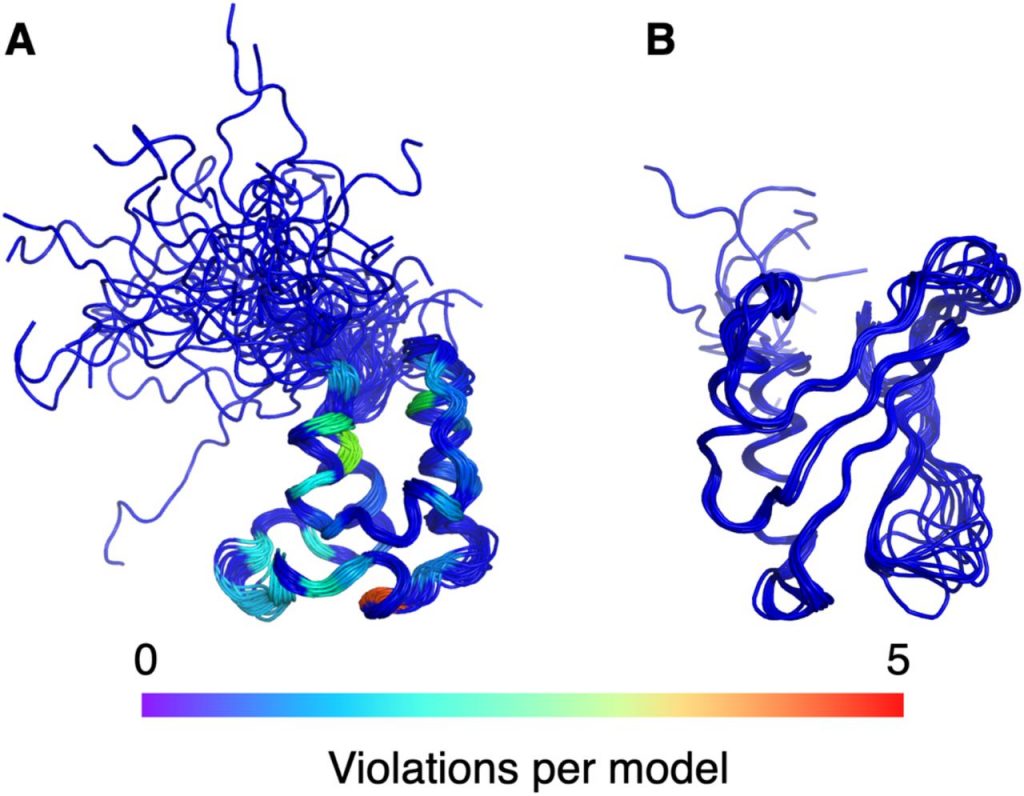
Baskaran H, Ploskon E, Tejero R, Yokochi M, Harrus D, Liang Y, …Montelione GT*, Vuister GW*, Young JY*. Restraint validation of biomolecular structures determined by NMR in the Protein Data Bank. Structure (Cell Press) Preprint.
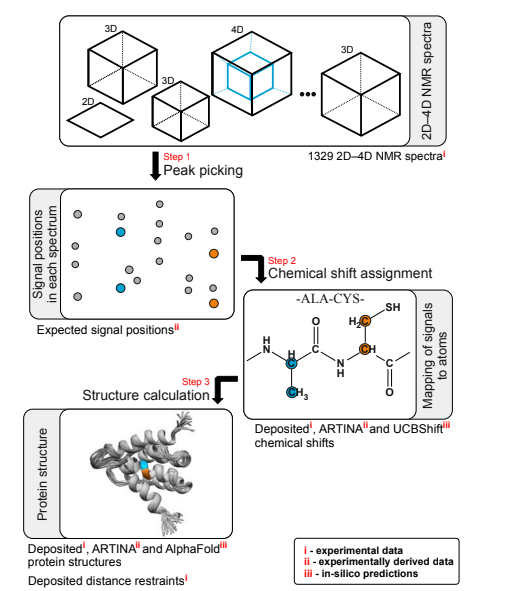
Klukowski P, Damberger FF, Allain FH, Iwai H, Kadavath H, Ramelot TA, Montelione GT, Riek R, Güntert P. The 100-protein NMR spectra dataset: A resource for biomolecular NMR data analysis. Scientific Data. 2024 Jan 4;11(1):30.
Gutsche I, Montelione GT. Editorial overview: Biophysical methods: Multiple structures of proteins underpin their biological functions. Curr Opin Struct Biol. 2024 Jan 12;84:102762. doi: 10.1016/j.sbi.2023.102762. Epub ahead of print. PMID: 38217897.