2019
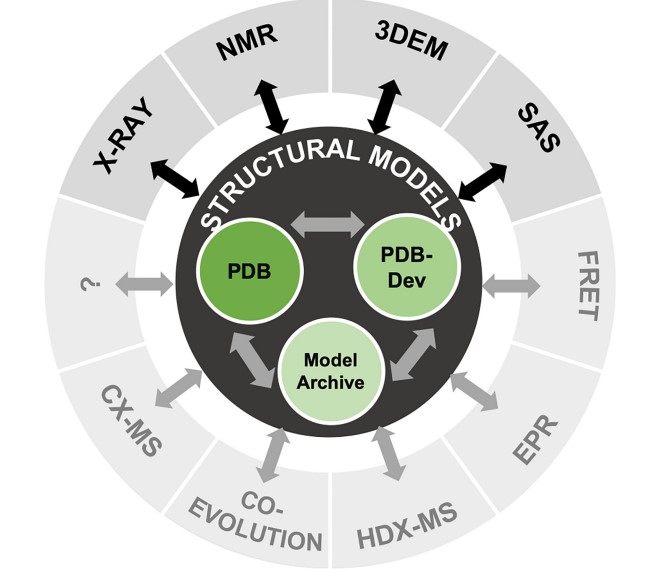
Berman HM, Adams PD, Bonvin AA, Burley SK, Carragher B, Chiu W, DiMaio F, Ferrin TE, Gabanyi MJ, Goddard TD, Griffin PR, Haas J, Hanke CA, Hoch JC, Hummer G, Kurisu G, Lawson CL, Leitner A, Markley JL, Meiler J, Montelione GT, Phillips GN Jr, Prisner T, Rappsilber J, Schriemer DC, Schwede T, Seidel CAM, Strutzenberg TS, Svergun DI, Tajkhorshid E, Trewhella J, Vallat B, Velankar S, Vuister GW, Webb B, Westbrook JD, White KL, Sali A. Federating Structural Models and Data: Outcomes from A Workshop on Archiving Integrative Structures. Structure. 2019 Dec 3;27(12):1745-1759.
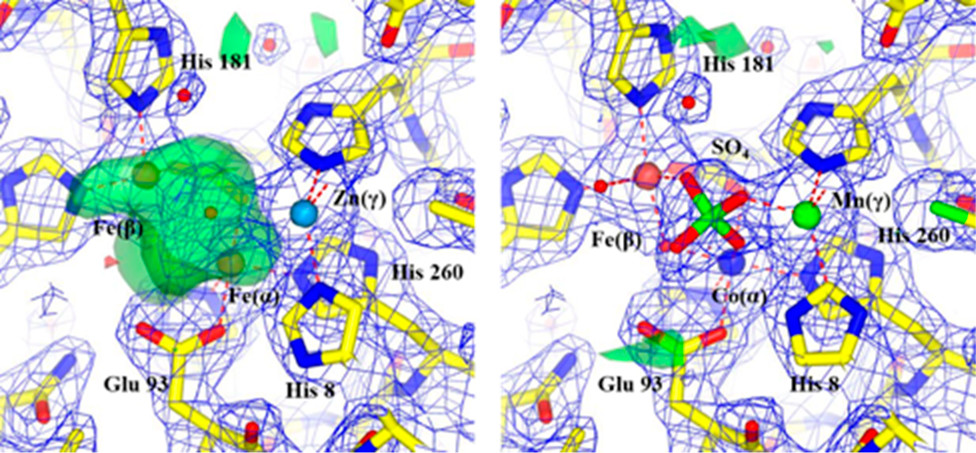
Grime GW, Zeldin OB, Snell ME, Lowe ED, Hunt JF, Montelione GT, Tong L, Snell EH, Garman EF. High-throughput PIXE as an essential quantitative assay for accurate metalloprotein structural analysis: development and application. Journal of the American Chemical Society. 2019 Dec 3;142(1):185-97.
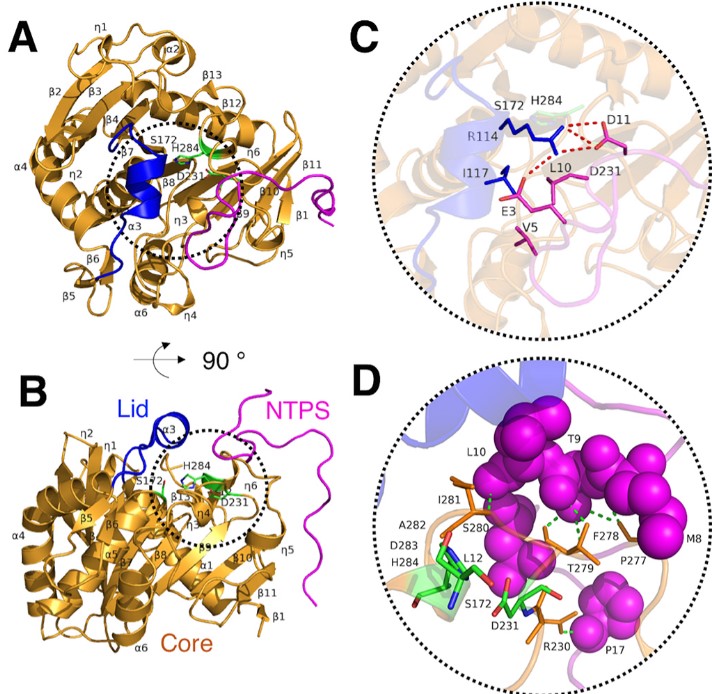
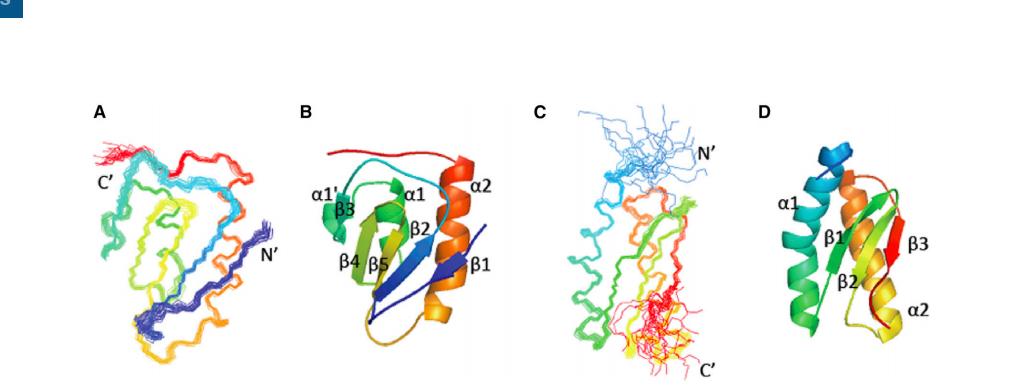
Zhang M, Yu XW, Xu Y, Guo RT, Swapna GVT, Szyperski T, Hunt JF, Montelione GT. Structural Basis by Which the N-Terminal Polypeptide Segment of Rhizopus chinensis Lipase Regulates Its Substrate Binding Affinity. Biochemistry. 2019 Sep 24;58(38):3943-3954.
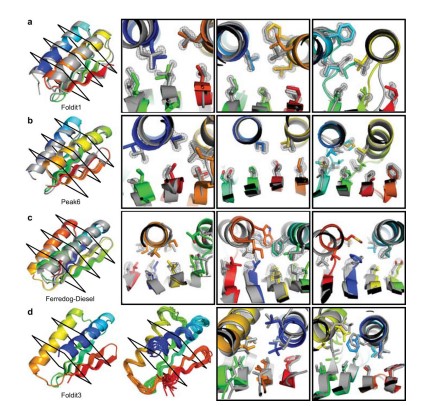
Koepnick B, Flatten J, Husain T, Ford A, Silva DA, Bick MJ, Bauer A, Liu G, Ishida Y, Boykov A, Estep RD, Kleinfelter S, Nørgård-Solano T, Wei L, Players F, Montelione GT, DiMaio F, Popović Z, Khatib F, Cooper S, Baker D. De novo protein design by citizen scientists. Nature. 2019 Jun;570(7761):390-394.
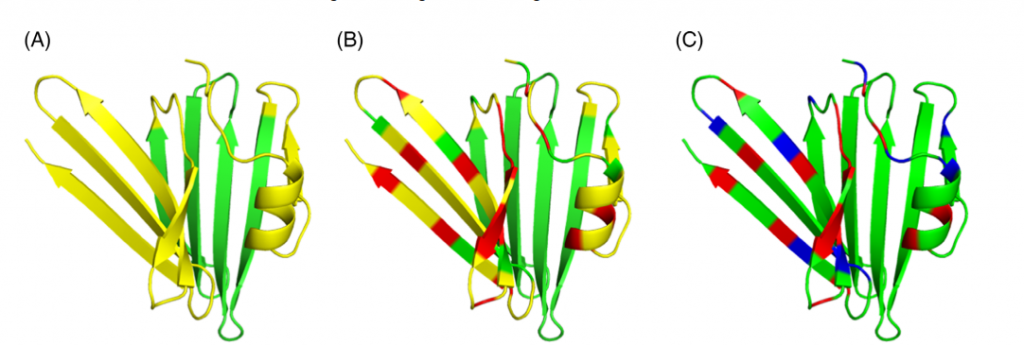
Sala, D., Huang, Y. J., Cole, C. A., Snyder, D. A., Liu, G., Ishida, Y., Swapna, G. V. T., Brock, K. P., Sander, C., Fidelis, K., Kryshtafovych, A., Inouye, M., Tejero, R., Valafar, H., Rosato, A., & Montelione, G. T. (2019). Protein structure prediction assisted with sparse NMR data in CASP13. Proteins, 87(12), 1315–1332.
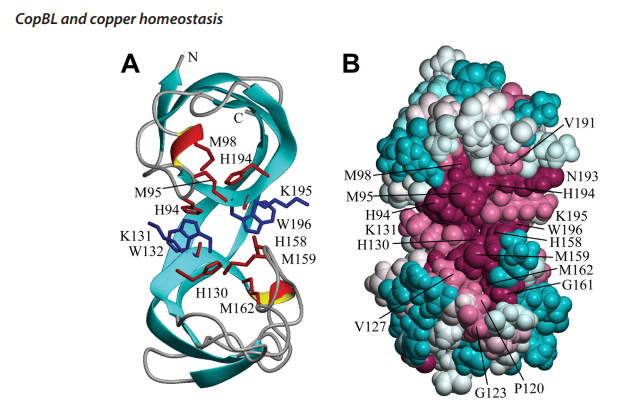
Rosario-Cruz, Z., Eletsky, A., Daigham, N. S., Al-Tameemi, H., Swapna, G. V. T., Kahn, P. C., Szyperski, T., Montelione, G. T., & Boyd, J. M. (2019). The copBL operon protects Staphylococcus aureus from copper toxicity: CopL is an extracellular membrane-associated copper-binding protein. The Journal of Biological Chemistry, 294(11), 4027–4044.
2018
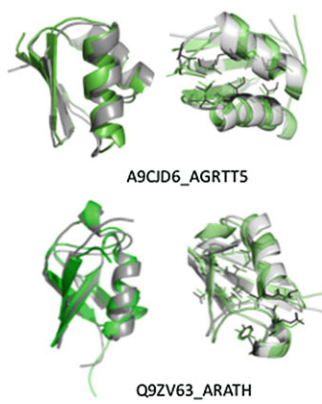
Huang YJ, Brock KP, Sander C, Marks DS, Montelione GT. A hybrid approach for protein structure determination combining sparse NMR with evolutionary coupling sequence data. Integrative structural biology with hybrid methods. 2018:153-69.
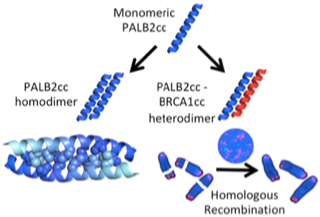
Song F, Li M, Liu G, Swapna GV, Daigham NS, Xia B, Montelione GT, Bunting SF. Antiparallel coiled-coil interactions mediate the homodimerization of the DNA damage-repair protein PALB2. Biochemistry. 2018 Oct 5;57(47):6581-91.
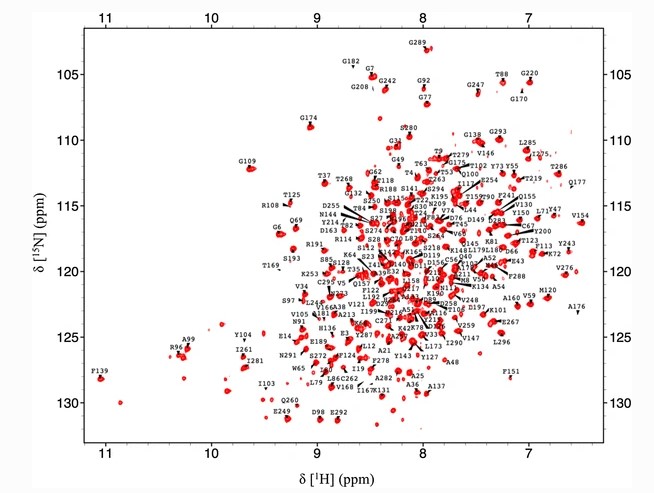
Zhang M; Yu X.-W; Swapna GVT; Liu G; Xiao R; Xu Y; Montelione GT. Backbone and Ile-δ 1, Leu, Val methyl 1H, 15N, and 13C, chemical shift assignments for Rhizopus chinensis lipase. Biomol NMR Assign 2018, 12: 63 – 68.
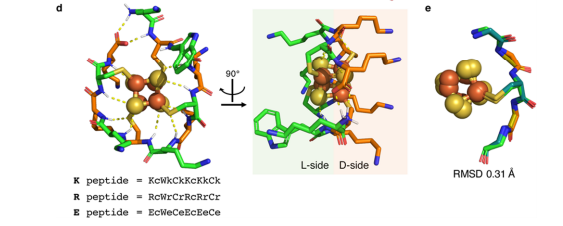
Kim J.D; Pike DH; Tyryshkin AM; Swapna GVT; Raanan H; Montelione GT; Nanda V; Falkowski PG. Minimal Heterochiral de Novo Designed 4Fe−4S Binding Peptide Capable of Robust Electron Transfer. J Amer Chem Soc. 2018, 140: 11210 – 11213.
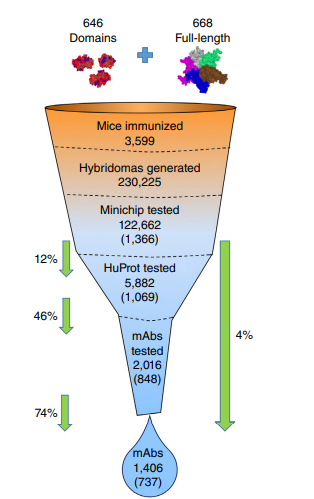
Venkataraman A; Yang K; Irizarry J; Mackiewicz M; Mita P; Kuang Z; Xue L.; Ghosh D; Liu S; Ramos P; Hu S; Bayron D; Keegan S; Saul R. Colantonio S; Zhang H; Behn FP; Song G; Albino E; Asencio L; Ramos L; Lugo L; Morell G; Rivera J; Ruiz K; Almodovar R; Nazario L; Murphy K; Vargas I; Rivera-Pacheco ZA; Rosa C; Vargas M; McDade J; Clark BS; Yoo S; Khambadkone SG; de Melo J; Stevanovic M; Jiang L; Li Y; Yap WY; Jones B; Tandon A; Campbell E; Montelione GT; Anderson S; Myers RM; Boeke JD; Fenyö D; Whiteley G; Bader JS; Pino I; Eichinger DJ; Zhu H; Blackshaw S. A toolbox of immunoprecipitation-grade monoclonal antibodies to human transcription factors. Nature Methods, 2018, 15: 303 – 338.
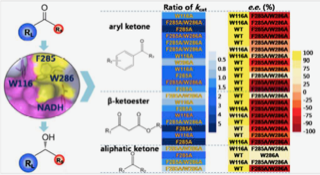
Nie Y, Wang S, Xu Y, Luo S, Zhao YL, Xiao R, Montelione GT, Hunt JF, Szyperski T. Enzyme engineering based on X-ray structures and kinetic profiling of substrate libraries: alcohol dehydrogenases for stereospecific synthesis of a broad range of chiral alcohols. ACS Catalysis. 2018 Apr 4;8(6):5145-52.
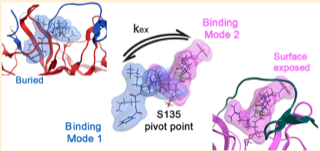
Gibbs AC, Steele R, Liu G, Tounge BA, Montelione GT. Inhibitor bound dengue NS2B-NS3pro reveals multiple dynamic binding modes. Biochemistry. 2018 Feb 15;57(10):1591-602.
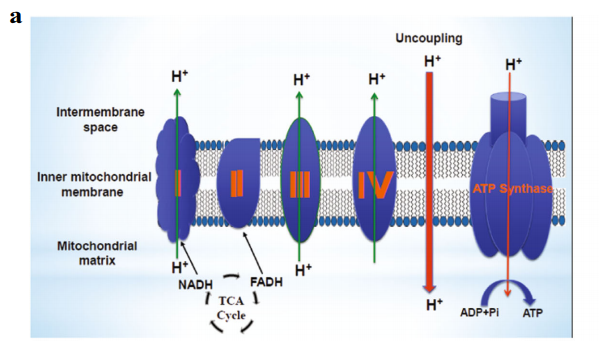
Alasadi A, Chen M, Swapna GV, Tao H, Guo J, Collantes J, Fadhil N, Montelione GT, Jin S. Effect of mitochondrial uncouplers niclosamide ethanolamine (NEN) and oxyclozanide on hepatic metastasis of colon cancer. Cell death & disease. 2018 Feb 13;9(2):215.
2017
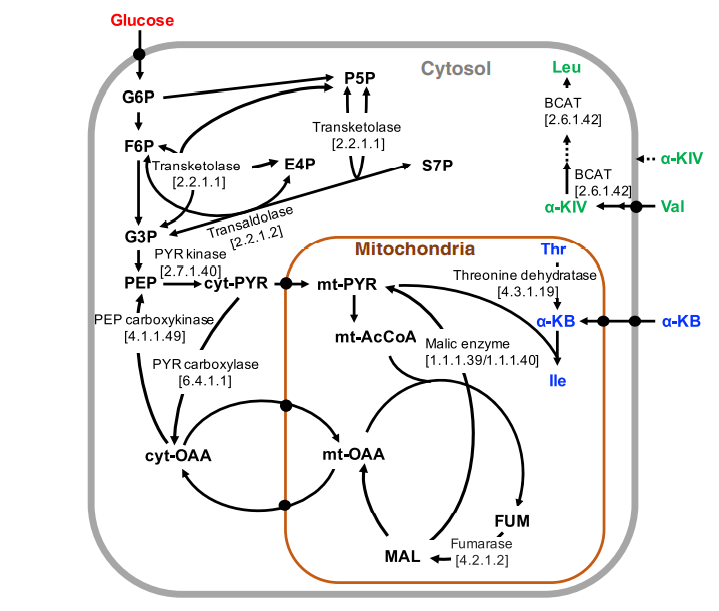
Zhang M, Yu XW, Xu Y, Jouhten P, Swapna GV, Glaser RW, Hunt JF, Montelione GT, Maaheimo H, Szyperski T. 13C metabolic flux profiling of Pichia pastoris grown in aerobic batch cultures on glucose revealed high relative anabolic use of TCA cycle and limited incorporation of provided precursors of branched‐chain amino acids. The FEBS journal. 2017 Sep;284(18):3100-13.
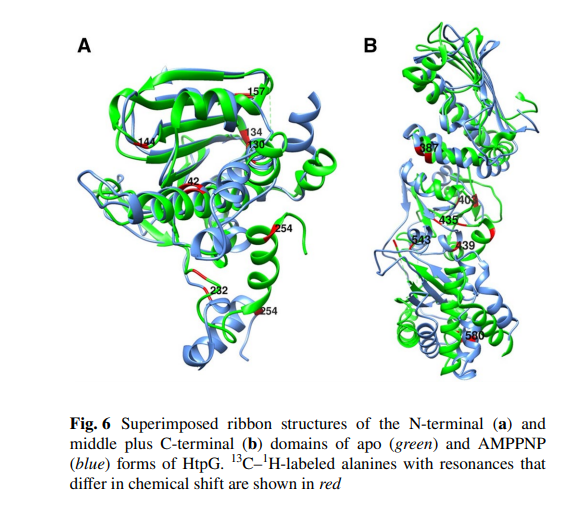
Pederson K, Chalmers GR, Gao Q, Elnatan D, Ramelot TA, Ma LC, Montelione GT, Kennedy MA, Agard DA, Prestegard JH. NMR characterization of HtpG, the E. coli Hsp90, using sparse labeling with 13 C-methyl alanine. Journal of biomolecular NMR. 2017 Jul;68:225-36.
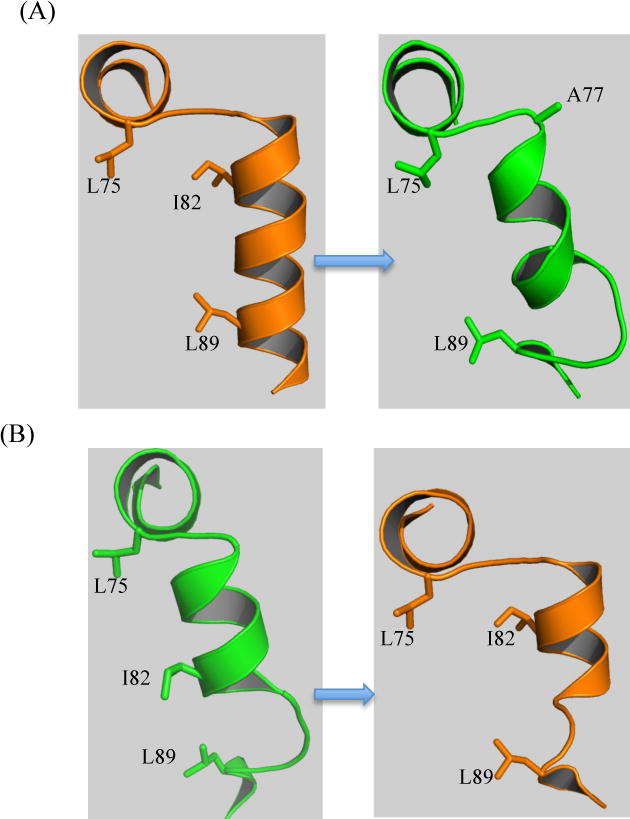
Harish B, Swapna GV, Kornhaber GJ, Montelione GT, Carey J. Multiple helical conformations of the helix‐turn‐helix region revealed by NOE‐restrained MD simulations of tryptophan aporepressor, TrpR. Proteins: Structure, Function, and Bioinformatics. 2017 Apr;85(4):731-40.
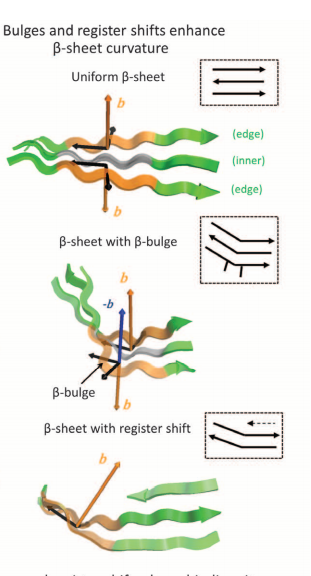
Marcos E, Basanta B, Chidyausiku TM, Tang Y, Oberdorfer G, Liu G, Swapna GV, Guan R, Silva DA, Dou J, Pereira JH, Xiao R, Sankaran B, Zwart PH, Montelione GT, Baker D. Principles for designing proteins with cavities formed by curved β sheets. Science. 2017 Jan 13;355(6321):201-206.
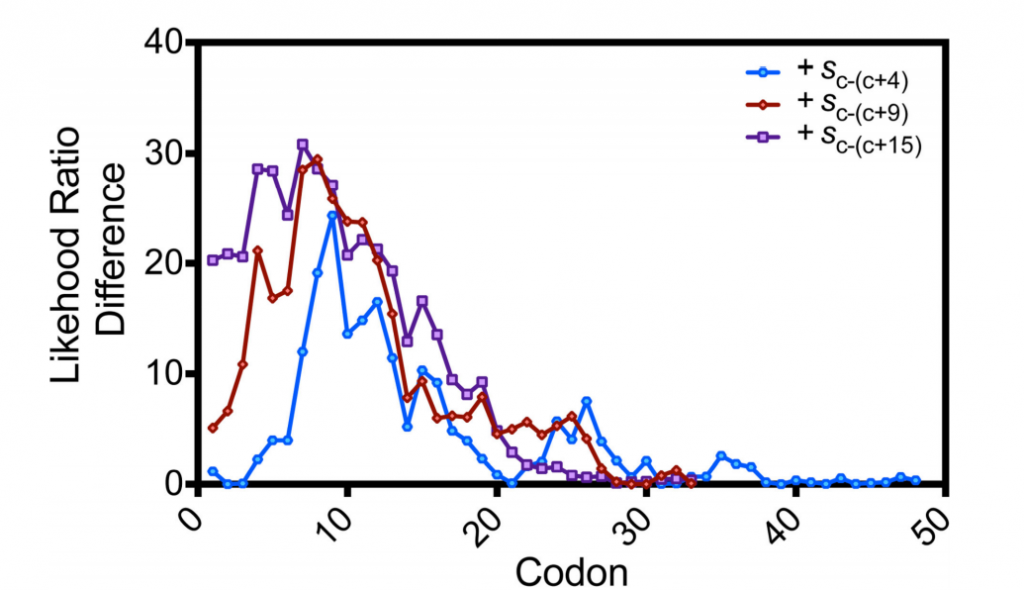
Boël G, Letso R, Neely H, Price WN, Wong KH, Su M, Luff J, Valecha M, Everett JK, Acton TB, Xiao R, Montelione GT, Aalberts DP, Hunt JF. Codon influence on protein expression in E. coli correlates with mRNA levels. Nature. 2016 Jan 21;529(7586):358-363.
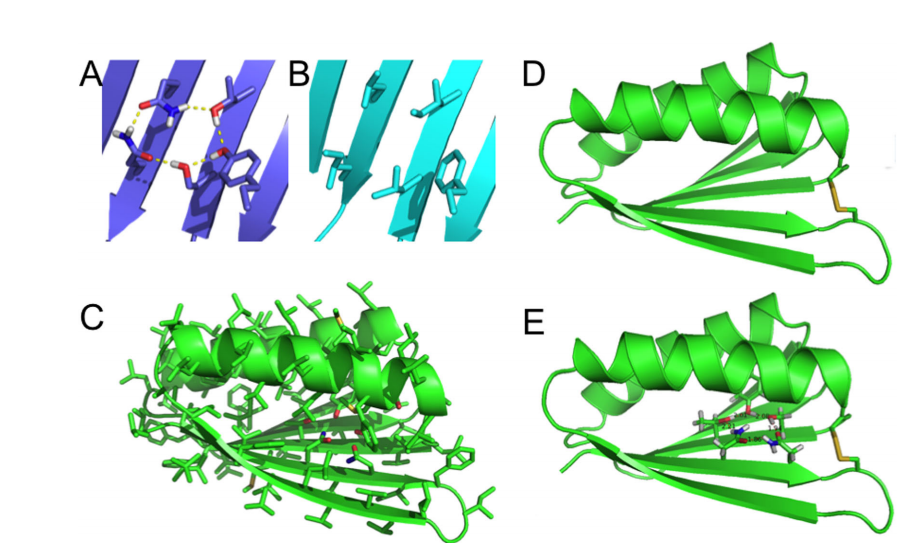
Basanta B; Kui B; Chan K; Barth P; King T; Hinshaw JR; Sosnick TR; Liu G; Everett J; Xiao R; Montelione GT; Baker D. Protein Science. 2016, 25: 1299 – 1307. Introduction of a polar core into the de novo designed protein Top7. suppl. material. PMC4918430. Pubmed.
Adams PD; Aertgeerts K; Bauer C; Bell JA; Berman HM; Bhat TN; Blaney J; Bolton E; Bricogne G; Brown D; Burley SK; Case DA; Clark KL; Darden T; Emsley P; Feher V; Feng Z; Groom CR; Harris SF; Hendle J; Holder T; Joachimiak A; Kleywegt G; Krojer T; Marcotrigiano J; Mark AE; Markley JL; Miller M; Minor W; Montelione GT; Murshudov G; Nakagawa A; Nakamura H; Nichols A; Nicklaus M; Nolte R; Padyana AK; Peishoff CE; Pieniazek S; Read RJ; Shao C; Sheriff S; Smart O; Soisson S; Spurlino J; Stouch T; Svobodova R; Tempel W; Terwilliger T; Tronrud D; Velankar S; Ward S; Warren G; Westbrook JD; Williams P; Yang H; Young J. Structure (Cell Press). 2016, 24: 502 – 508. Outcome of the first wwPDB/CCDC/D3R ligand validation workshop. suppl. material. PMC5070601. Pubmed.
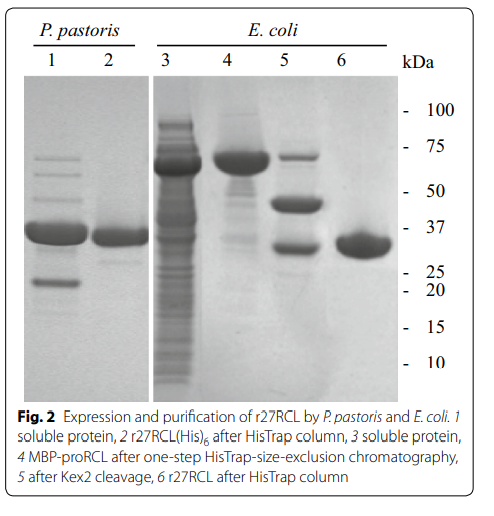
Zhang M, Yu XW, Swapna GV, Xiao R, Zheng H, Sha C, Xu Y, Montelione GT. Efficient production of 2 H, 13 C, 15 N-enriched industrial enzyme Rhizopus chinensis lipase with native disulfide bonds. Microbial Cell Factories. 2016 Dec;15:1-2.
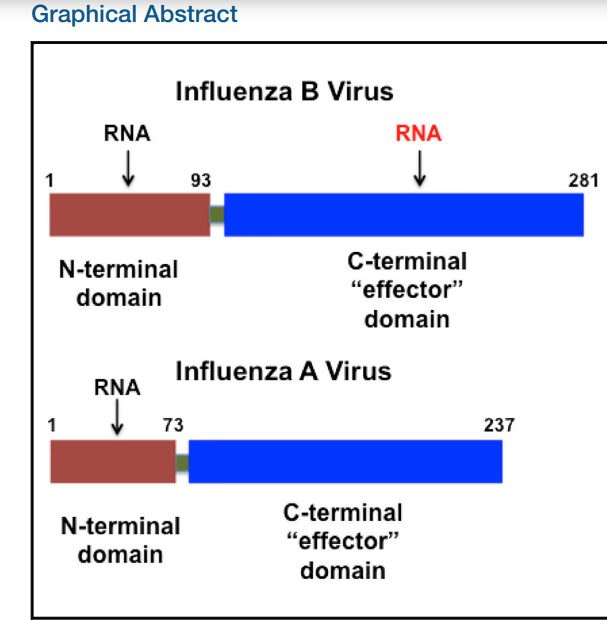
Ma L-C; Guan R; Hamilton K; Aramini J; Mao L; Wang S; Krug RM; Montelione GT. Structure (Cell Press). 2016, 24: 1562 – 1572. A second RNA-binding site in the NS1 protein of influenza B virus. suppl. material 1 suppl. material 2. PMC5014651. Pubmed.
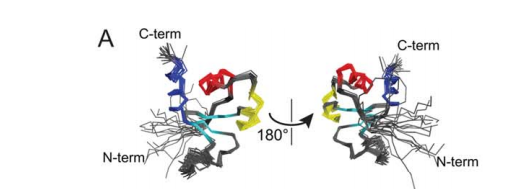
Sachleben JR; Adhikari AN; Gawlak G; Hoey RJ; Liu G; Joachimiak A; Montelione GT; Sosnick TR; Koide S. Protein Science. 2016, 26: 208 – 217. Aromatic Claw: A new fold with high aromatic content that evades structural prediction. suppl. material. PMC5275723. Pubmed.
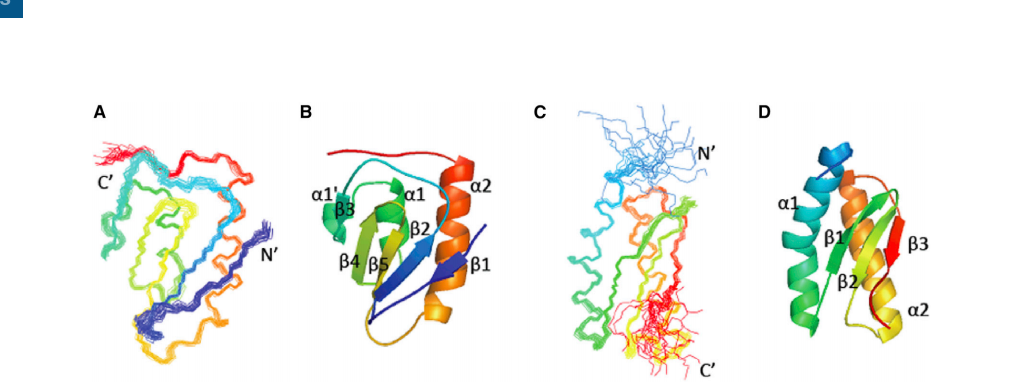
Cai K; Liu G; Frederick RO; Xiao R; Montelione GT; Markley JL. Structure (Cell Press). 2016, 24: 2080 – 2091. Structural/functional properties of human NFU1, an intermediate [4Fe-4S] carrier in human mitochondrial iron-sulfur cluster biogenesis. suppl. material 1. suppl. material 2. PMC5166578. Pubmed.
2015
1. Basanta B; Kui B; Chan K; Barth P; King T; Hinshaw JR; Sosnick TR; Liu G; Everett J; Xiao R; Montelione GT; Baker D. Protein Science. 2016, 25: 1299 – 1307. Introduction of a polar core into the de novo designed protein Top7. suppl. material. PMC4918430. Pubmed.
2. Adams PD; Aertgeerts K; Bauer C; Bell JA; Berman HM; Bhat TN; Blaney J; Bolton E; Bricogne G; Brown D; Burley SK; Case DA; Clark KL; Darden T; Emsley P; Feher V; Feng Z; Groom CR; Harris SF; Hendle J; Holder T; Joachimiak A; Kleywegt G; Krojer T; Marcotrigiano J; Mark AE; Markley JL; Miller M; Minor W; Montelione GT; Murshudov G; Nakagawa A; Nakamura H; Nichols A; Nicklaus M; Nolte R; Padyana AK; Peishoff CE; Pieniazek S; Read RJ; Shao C; Sheriff S; Smart O; Soisson S; Spurlino J; Stouch T; Svobodova R; Tempel W; Terwilliger T; Tronrud D; Velankar S; Ward S; Warren G; Westbrook JD; Williams P; Yang H; Young J. Structure (Cell Press). 2016, 24: 502 – 508. Outcome of the first wwPDB/CCDC/D3R ligand validation workshop. suppl. material. PMC5070601. Pubmed.
3. Zhang M; Yu XW; Swapna GVT; Xiao R; Zheng H; Sha C; Xu Y; Montelione GT. Microbial Cell Factories. 2016, 15: 123. Efficient production of 2H, 13C, 15N-enriched proteins with native disulfide bonds. Application to the enzyme Rhizopus chinensis lipase. PMC494443. Pubmed.
4. Ma L-C; Guan R; Hamilton K; Aramini J; Mao L; Wang S; Krug RM; Montelione GT. Structure (Cell Press). 2016, 24: 1562 – 1572. A second RNA-binding site in the NS1 protein of influenza B virus. suppl. material 1 suppl. material 2. PMC5014651. Pubmed.

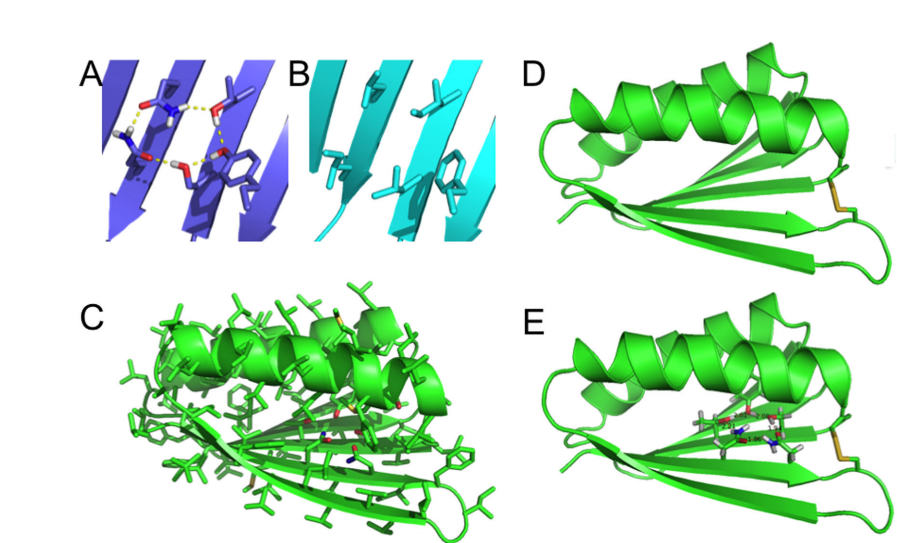
5. Sachleben JR; Adhikari AN; Gawlak G; Hoey RJ; Liu G; Joachimiak A; Montelione GT; Sosnick TR; Koide S. Protein Science. 2016, 26: 208 – 217. Aromatic Claw: A new fold with high aromatic content that evades structural prediction. suppl. material. PMC5275723. Pubmed.
6. Cai K; Liu G; Frederick RO; Xiao R; Montelione GT; Markley JL. Structure (Cell Press). 2016, 24: 2080 – 2091. Structural/functional properties of human NFU1, an intermediate [4Fe-4S] carrier in human mitochondrial iron-sulfur cluster biogenesis. suppl. material 1. suppl. material 2. PMC5166578. Pubmed.