2015
1. Parmeggiani F; Huang P-S; Vorobiev S; Xiao R; Park K; Caprari S; Su M; Jayaraman S; Mao L; Janjua H; Montelione GT; Hunt JF; Baker D. J Mol Biol. 2015, 427: 563 – 575. Beyond consensus: a general computational approach for repeat protein design. suppl. material 1 suppl. material 2 MC4303030. Pubmed.
2. Rossi P; Shi L; Liu G; Barbieri CM; Lee H-W; Grant TD; Luft JR; Xiao R; Acton TB; Snell EH; Montelione GT; Baker D; Lange OF; Sgourakis NG. PROTEINS: Struc. Funct. Bioinformatics. 2015, 83: 309 – 317. A hybrid NMR/SAXS-based approach for discriminating oligomeric protein interfaces using Rosetta. suppl. material. PMC5061451. Pubmed.
3. Luft JR; Wolfley JR; Franks EC; Lauricella AM; Gualtieri EJ; Snell EH; Xiao R; Everett JK; Montelione GT. Structural Dynamics. 2015, 2: 041710. The detection and subsequent volume optimization of biological nanocrystals. PMC4711624.
4. Aiyer S; Rossi P; Malani N; Schneider WM; Chanda A; Bushman FD; Montelione GT; Roth MJ. Nucleic Acids Research. 2015, 43: 5647 – 5663. Structural and sequencing analysis of local target DNA recognition by MLV integrase. PMC4477651. Pubmed.
5. Sali A; Berman HM; Schwede T; Trewhella J; Kleywegt G; Burley SK; Markley J; Nakamura H; Adams P; Bonvin AMJJ.; Chiu W; Dal Peraro M; Di Maio F; Ferrin TE; Grünewald K; Gutmanas A; Henderson R; Hummer G; Iwasaki K; Johnson G; Lawson KL; Meiler J; Marti-Renom MA; Montelione GT; Nilges M; Nussinov R; Patwardhan A; Rappsilber J; Read RJ; Saibil H; Schröder GF; Schwieters C; Seidel CAM; Svergun D; Topf M; Ulrich EL; Velankar S; Westbrook JD. Structure (Cell Press). 2015, 23: 1156 – 1167. Outcome of the first wwPDB hybrid / integrative methods task force workshop. PMC4933300. Pubmed.

6. Gutmanas A; Adams PD; Bardiaux B; Berman HM; Case DA; Fogh RH; Güntert P; Hendrickx PMS; Herrmann T; Kleywegt GJ; Kobayashi N; Lange OF; Markley JL; Montelione GT; Nilges M; Ragan TJ; Schwieters, CD; Tejero R; Ulrich E; Velankar S; Vranken WF; Wedell JR; Westbrook J; Wishart DS; Vuister GW Nature Struct Mol Biol. 2015, 22: 433 – 434. NMR Exchange Format: a unified and open standard for representation of NMR restraint data. PMC4546829. Pubmed.
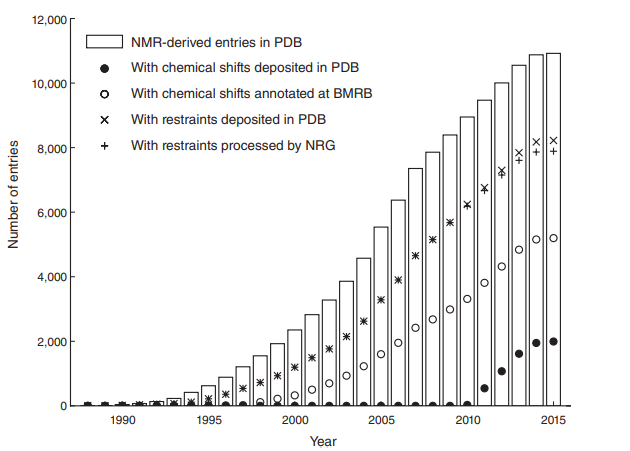
7. Ragan TJ; Fogh RH; Tejero R; Vranken W; Montelione GT; Rosato A; Vuister GW. J Biomol NMR. 2015, 62: 527 – 540. Analysis of the structural quality of the CASD-NMR 2013 entries. suppl. material 1 suppl. material 2. PMC4569653. Pubmed.
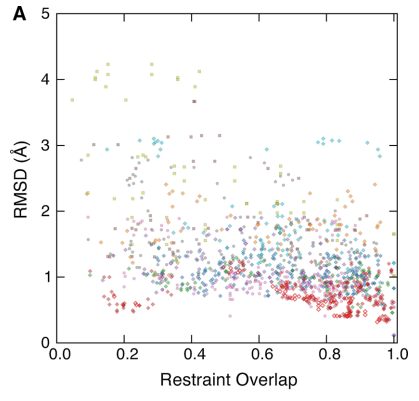
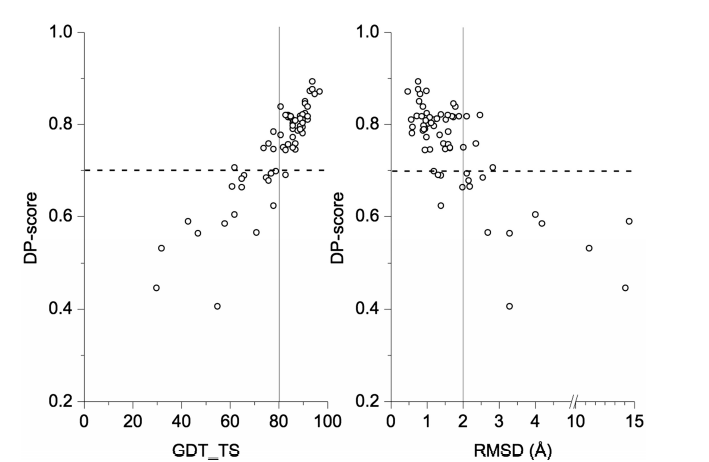
8. Huang YJ; Mao B; Xu F; Montelione GT. J Biomol NMR. 2015, 62: 439 – 451. Guiding automated NMR structure determination using a global optimization metric, the NMR DP score. PMC4943320. Pubmed.
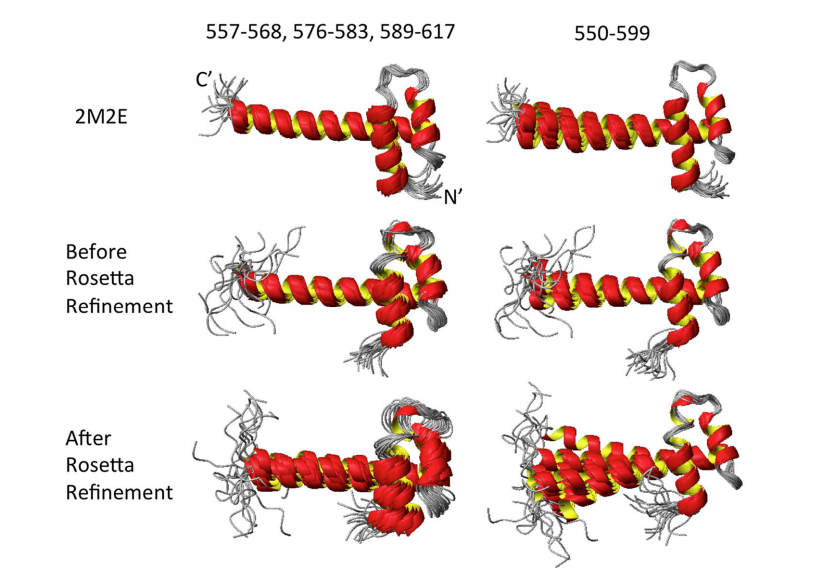
9. Rosato A; Vranken W; Fogh RH; Ragan TJ; Tejero R; Pederson K; Lee HW; Prestegard J; Yee A; Wu B; Lemak A; Houliston S; Arrowsmith C; Kennedy M; Acton TB; Liu G; Xiao R; Montelione GT; Vuister GW. J Biomol NMR. 2015, 62: 413 – 424. The second round of critical assessment of automated structure determination of proteins by NMR: CASD-NMR-2013. suppl. material. PMC4569658.

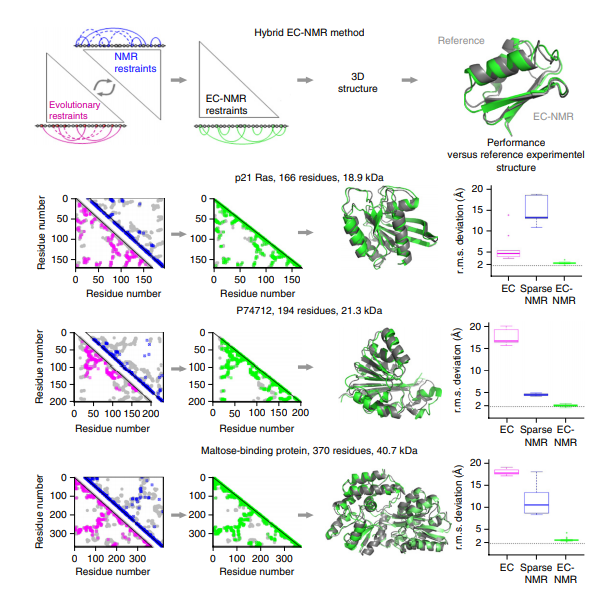
10. Tang Y; Huang YJ; Hopf TA; Sanders C; Marks DS; Montelione GT. Nature Methods. 2015, 12: 751 – 754. Protein structure determination by combining sparse NMR spectroscopy data with evolutionary couplings. suppl. material. PMC4521990. Pubmed.
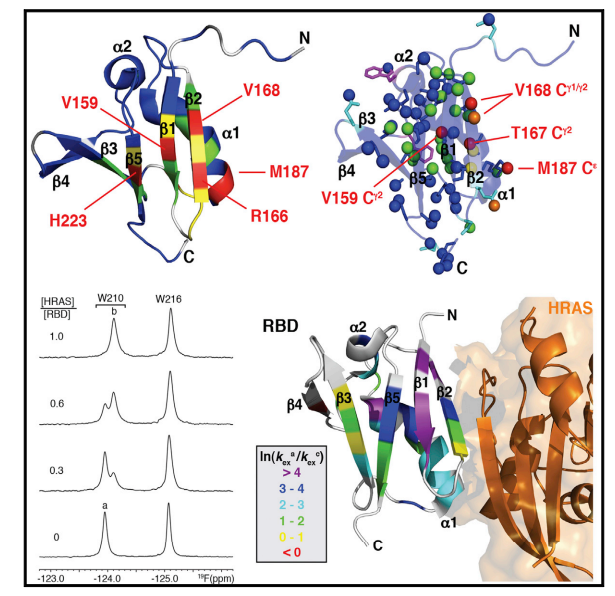
11. Aramini JM; Vorobiev SM; Tuberty LM; Janjua H; Campbell ET; Seethraman J; Su M; Huang YJ; Acton TB; Xiao R; Tong L; Montelione GT. Structure (Cell Press). 2015, 23: 1 – 12. The RAS- binding domain of human BRAF protein serine/threonine kinase exhibits allosteric conformational changes upon binding HRAS. suppl. material 1. suppl. material 2. PMC4963008. Pubmed.
12. Choi HW; Tian M; Song F; Venereau E; Preti A; Park SW; Hamilton K; Swapna GVT; Manohar M; Moreau M; Agresti A; Gorzanelli A; De Marchis F; Wang H; Antonyak M; Micikas RJ; Gentile DR; Cerione RA; Schroeder FC; Montelione GT; Bianchi ME; Klessig DF. Molecular Medicine. 2015, 21: 526 – 535. Aspirin’s active metabolite salicylic acid targets High Mobility 1 Group Box 1 (HMGB1) to modulate inflammatory responses. PMC4607614. Pubmed.
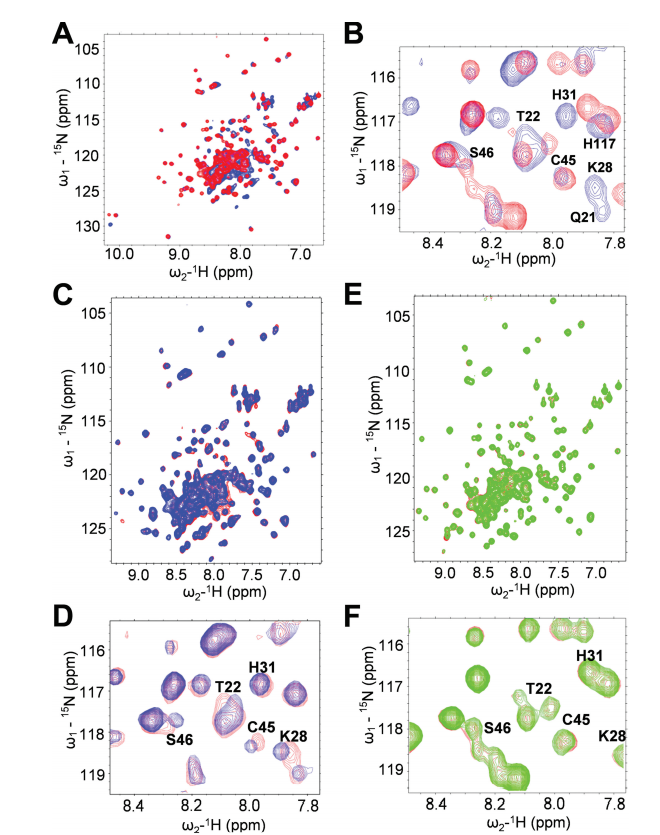
13. Everett JK; Tejero R; Murthy SBK; Acton TB; Aramini JM; Baran MC; Benach J; Cort JR; Eletsky A; Forouhar F; Guan R; Kuzin AP; Lee HW; Liu G; Mani R; Mao B; Mills JL; Montelione AF; Pederson K; Powers R; Ramelot T; Rossi P; Seetharaman J; Snyder D; Swapna GVT; Vorobiev SM; Wu Y; Xiao R; Yang Y; Arrowsmith CH; Hunt JF; Kennedy MA; Prestegard JH; Szyperski T; Tong L; Montelione GT. Protein Science. 2015, 25: 30 – 45. A community resource of experimental data for NMR – X-ray crystal structure pairs. suppl. material. PMC4815321.
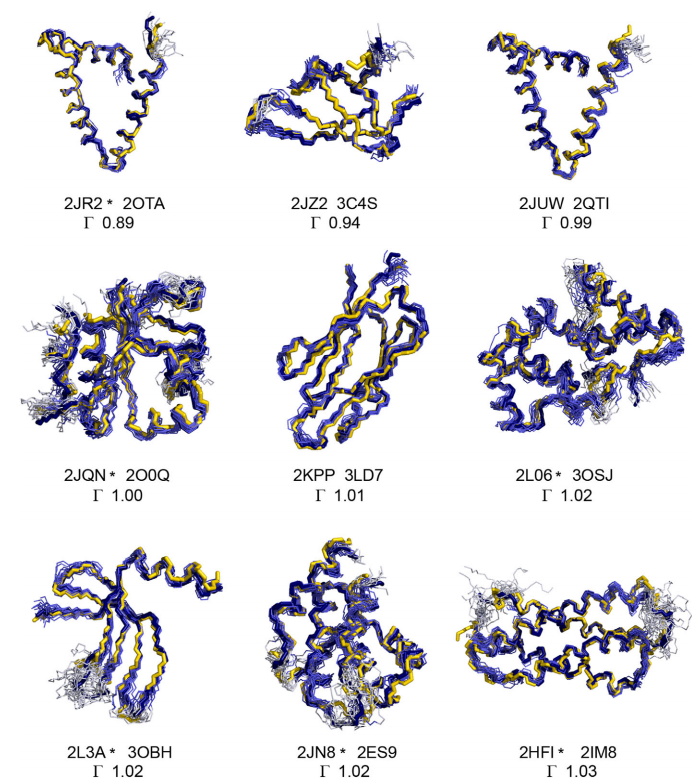
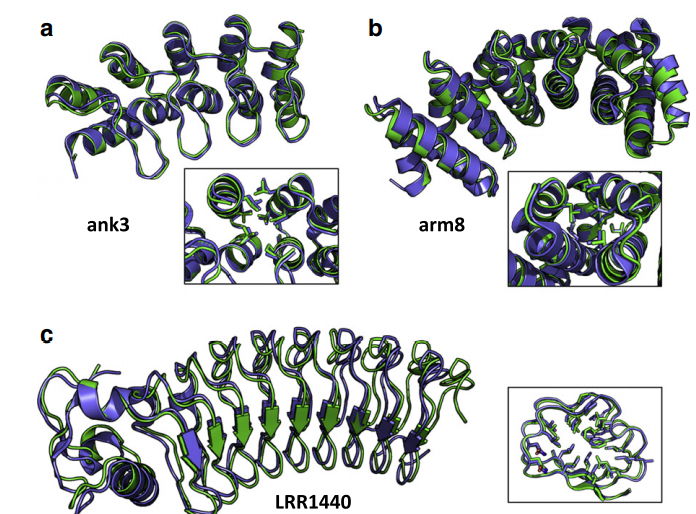
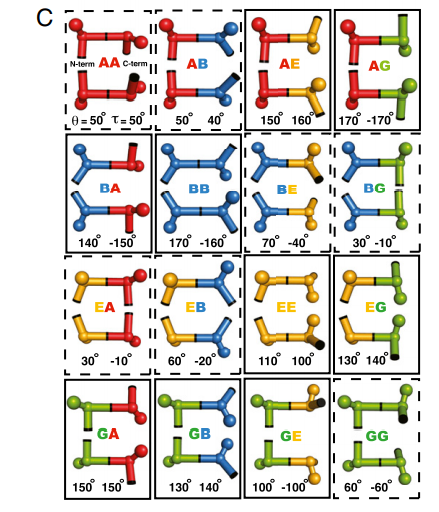
14. Lin YR; Koga N; Tatsumi-Koga R; Liu G; Clouser AF; Montelione GT; Baker D. Proc Natl Acad Sci USA. 2015, 112: e5478 – 5485. Control over overall shape and size in de novo design proteins. suppl. material. PMC4603489. Pubmed.
15. Wolf C; Siegel JB; Tinberg C; Camarca A; Gianfrani C; Paski S; Guan R; Montelione GT; Baker D; Pultz IS. J Amer Chem Soc 2015, 137: 13106 – 13113. Engineering of Kuma030: a gliadin peptidase that rapidly degrades immunogenic gliadin peptides in gastric conditions. suppl. material. PMC4958374. Pubmed.

16. King IC; Gleixner J; Doyle L; Kuzin A; Hunt JF; Xiao R; Montelione GT; Stoddard BL; DiMaio F; Baker D. eLIFE. 2015, 4: e11012. Precise assembly of complex beta sheet topologies from de novo design building blocks. suppl. material. PMC4737653. Pubmed.
2014
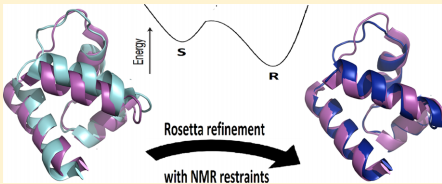
1. Mao B; Tejero R; Baker D; Montelione GT. J Amer Chem Soc. 2014, 136: 1893 – 1906. Protein NMR structures refined with Rosetta have higher accuracy relative to corresponding X-ray crystal structures. suppl material. PMC4129517. Pubmed.
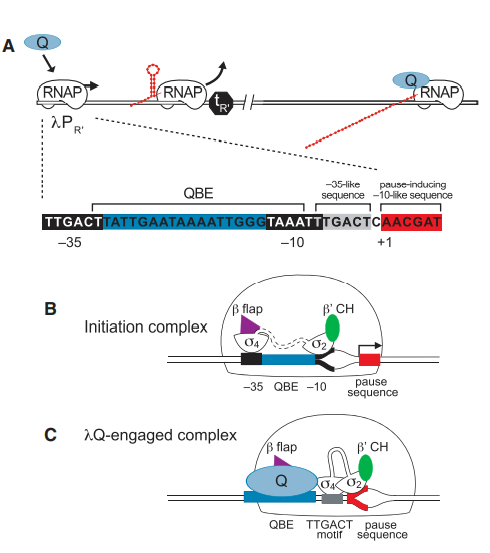
2. Vorobiev S; Gensler Y; Vahedian-Movahed H; Seetharaman J; Su M; Huang Y-P; Xiao R; Kornhaber G; Montelione GT; Tong L; Ebright RH; Nickels BE. Structure (Cell Press). 2014, 22: 488 – 495. Structure of the DNA-binding and RNA polymerase-binding region of transcription antitermination factor λQ. suppl. material. PMC3951671. Pubmed.
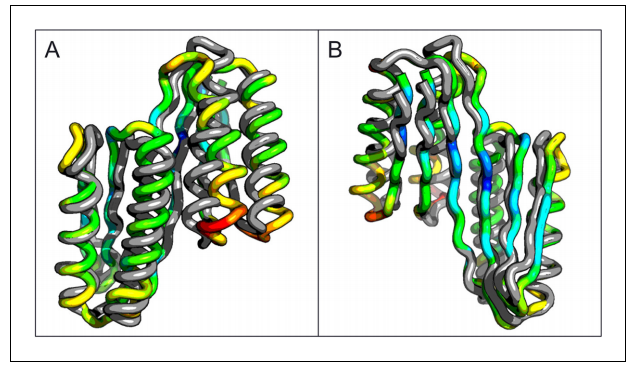
3. Srinivisan B; Forouhar F; Shukla A; Sampangi C; Kulkarni S; Abashidze M; Seetharaman J; Lew S; Mao L; Acton T; Xiao R; Everett J; Montelione GT; Tong L; Balaram H. FEBS Lett. 2014, 281: 1613 – 1628. Allosteric regulation and substrate activation in cytosolic nucleotidase II from Legionella pneumophila suppl. material 1 suppl. material 2. PMC3982195. Pubmed.
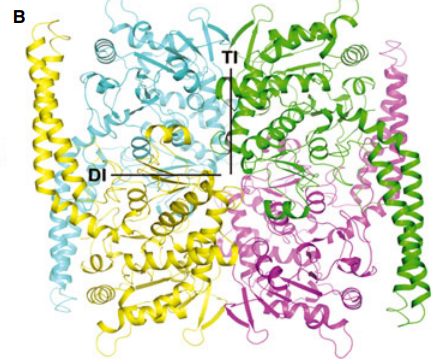
4. Stark JL; Mehla K; Chaika N; Acton TB; Xiao R; Singh PK; Montelione GT; Powers R. Biochemistry. 2014, 53: 1360 – 1372. The structure and function of DnaJ homolog subfamily A member 1 (DNAJA1) and its relationship to pancreatic cancer. PMC3985919. Pubmed.

5. Aramini JM; Hamilton K; Ma L-C; Swapna GVT; Leonard PG; Ladbury JE; Krug RM; Montelione GT. Structure (Cell Press). 2014, 22: 515 – 525. F NMR reveals multiple conformations at the dimer interface of the NS1 effector domain from influenza A virus. suppl. material. PMC4110948. Pubmed.
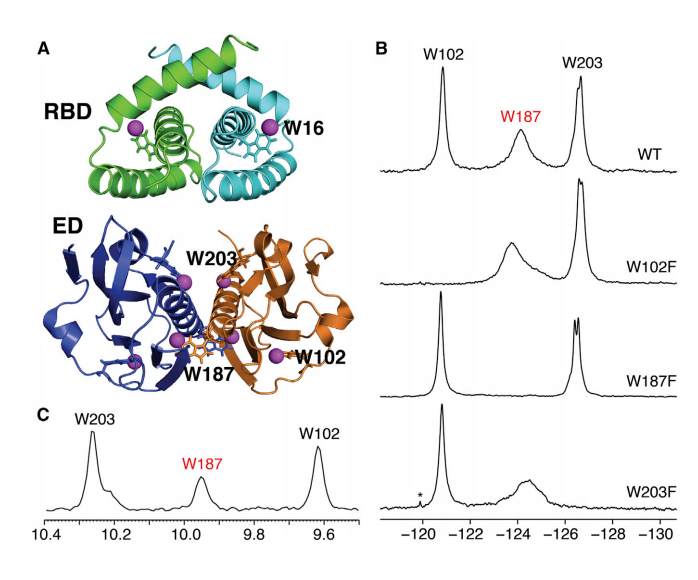
6. Aiyer S; Swapna GVT; Malani N; Aramini JM; Schneider WM; Plumb MR; Ghanem M; Larue RC; Sharma A; Studamire B; Kvaratskhelia M; Bushman FD; Montelione GT; Roth MJ. Nucleic Acids Research. 2014, 42: 5917 – 5928. Altering murine leukemia virus integration through disruption of the integrase and BET protein family interaction suppl. material. PMC4027182. Pubmed.
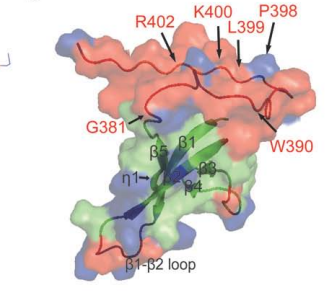
7. Pulavarti S; Eletsky A; Huang YJ; Acton TB; Xiao R; Everett JK; Montelione GT; Szyperski T. Biomol NMR Assign. 2014, 9: 135 – 138. Polypeptide backbone, Cb and methyl group resonance assignments of the 24 kDa plectin repeat domain 6 from human protein plectin. PMC4194182. Pubmed.
8. Xu X; Pulavarti SV; Eletsky A; Huang YJ; Acton TB; Xiao R; Everett JK; Montelione GT; Szyperski T. J Struct Funct Genomics. 2014, 15: 201 – 207. Solution NMR structures of homeodomains from human proteins ALX4, ZHX1, and CASP8AP2 contribute to the structural coverage of the Human Cancer Protein Interaction Network. suppl. material. PMC4239167. Pubmed.
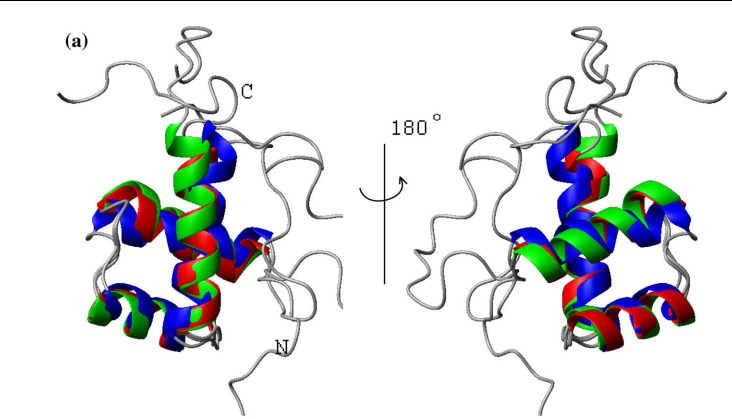
9. Sathyamoorthy B; Parish DM; Montelione GT; Xiao R; Szyperski T. Chem Physchem. 2014, 15: 1872 – 1879. Spatially selective heteronuclear multiple-quantum coherence spectroscopy for biomolecular NMR studies. suppl. material. PMC4121990. Pubmed.
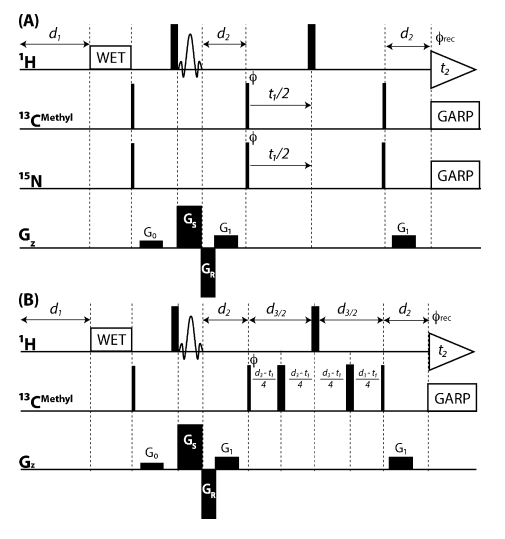
10. Bruno EB; Ruby AM; Luft JR; Grant TD; Seetharaman J; Montelione GT; Hunt JF; Snell EH. PLoS One. 2014, 9: e100782. Comparing chemistry to outcome: The development of a chemical distance metric coupled with clustering and hierarchal visualization applied to macromolecular crystallography. suppl. material. PMC4074061.
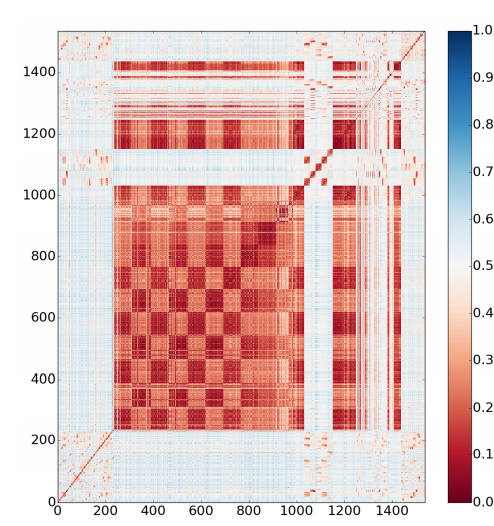
11. Pulavarti SV; Huang YJ; Pederson K; Acton TB; Xiao R; Everett JK; Prestegard JH; Montelione GT; Szyperski T. J Struct Funct Genomics. 2014, 15: 209 – 214. Solution NMR structures of immunoglobulin-like domains 7 and 12 from obscurin-like protein 1 contribute to the structural coverage of the human cancer protein interaction network. suppl. material. PMC4945113. Pubmed.
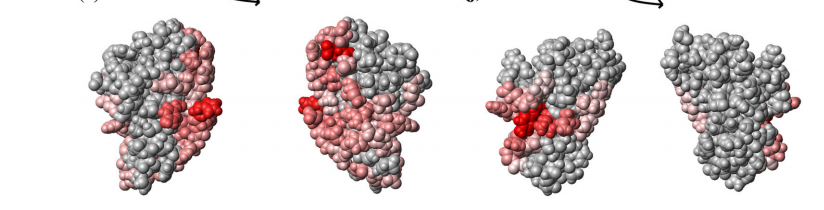
12. Eletsky A; Michalska K; Houliston S; Zhahttps://pubmed.ncbi.nlm.nih.gov/25010333/ng Q; Daily MD; Xu X; Cui H; Yee A; Lemak A; Wu B; Garcia M; Burnet MC; Meyer KM; Aryal UK; Sanchez O; Ansong C; Xiao R; Acton TB; Adkins JN; Montelione GT; Joachimiak A; Arrowsmith CH; Savchenko A; Szyperski T; Cort J.R. PLoS One. 2014, 9: e101787. Structural and functional characterization of DUF1471 domains of salmonella. suppl. material. PMC4092069. Pubmed.
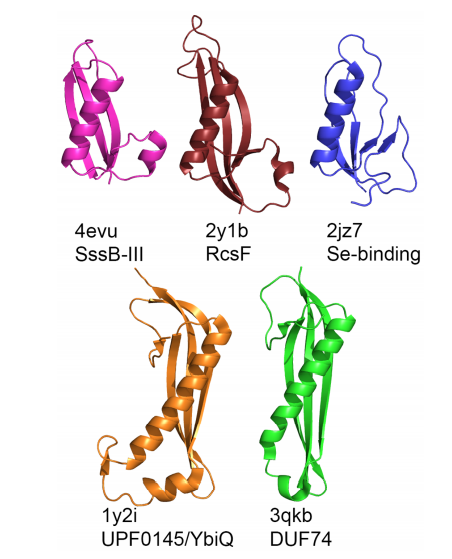
13. Elshahawi S; Ramelot T; Seetharaman J; Chen J; Singh S; Yang Y; Pederson K; Kharel M; Xiao R; Yennamalli R; Wang J; Tong L; Montelione G; Kennedy M; Bingman C; Phillips G; Thorson J. ACS Chemical Biology. 2014, 9: 2347 – 2358. Structure-guided functional characterization of enediyne self-sacrifice resistance proteins CalU16 and CalU19. suppl. material. PMC4201346. Pubmed.
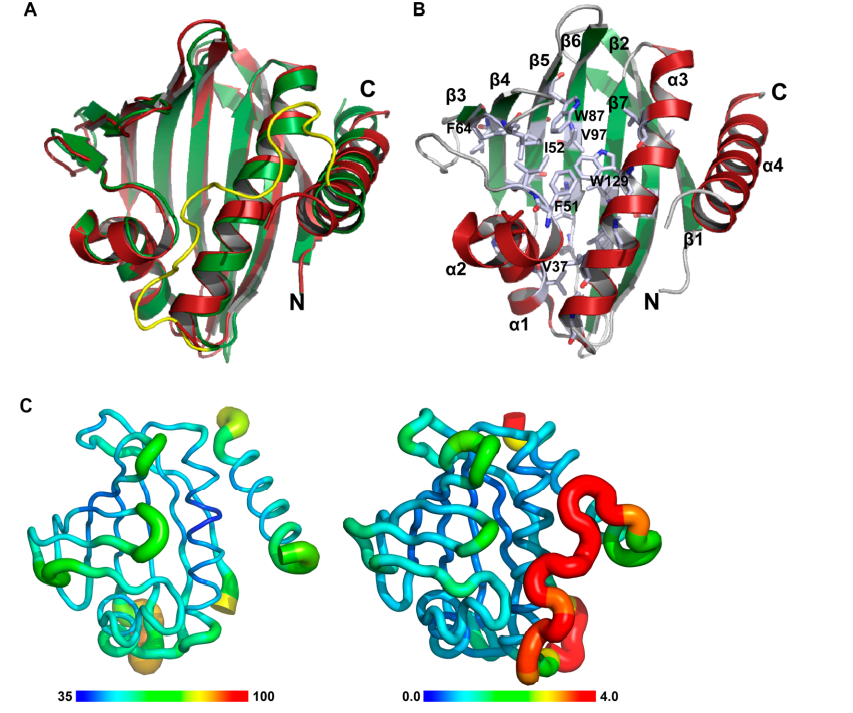
14. Yang Y; Ramelot TA; Lee HW; Xiao R; Everett JK; Montelione GT; Prestegard JH; Kennedy M. J Biomol NMR. 2014, 60: 189 – 195. Solution structure of the free Zα domain of human DLM-1 (ZBP1/DAI), a Z-DNA binding domain. PMC4527548. Pubmed.
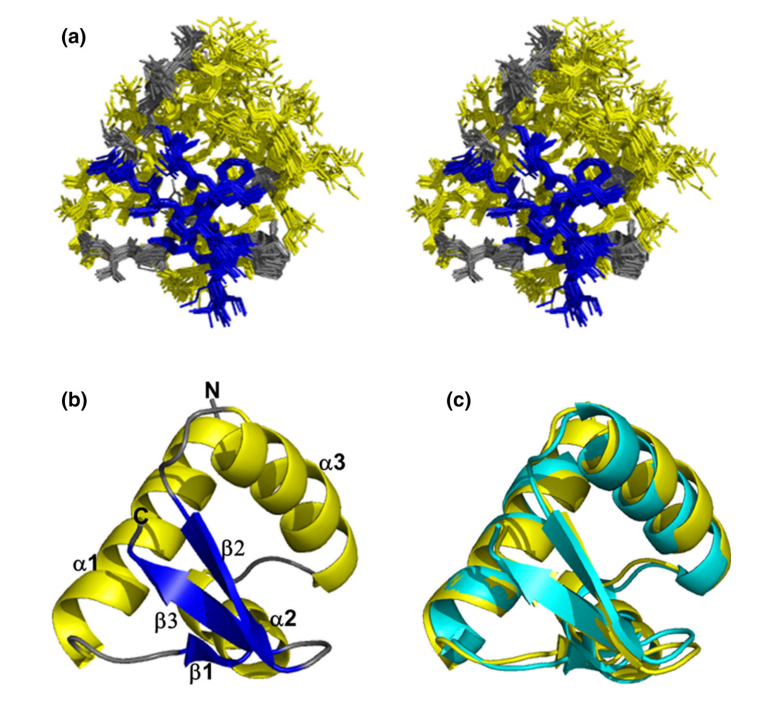
15. Yang Y; Ramelot TA; Lee HW; Xiao R; Everett, JK; Montelione GT; Prestegard JH; Kennedy M. J Biomol NMR. 2014, 60: 197 – 202. Solution structure of a C-terminal fragment (175-257) of CV_0373 protein from Chromobacterium violaceum adopts a winged helix-turn-helix (wHTH) fold. PMC4928572. Pubmed.
16. Bruno A; Ruby A; Luft J; Grant T; Seetharman J; Hunt J; Montelione G; Snell E. Acta Cryst. 2014, A70: C1145. Chemical clustering and visualization applied to macromolecular crystallography. suppl. material. PMC4074061. Pubmed.
17. Huang YJ; Mao B; Aramini J; Montelione GT. PROTEINS: Struct Funct Genomics. 2014, 82: Suppl 2: 43 – 56. Assessment of template based protein structure predictions in CASP10. suppl. material 1. suppl. material 2. suppl. material 3. suppl. material 4. PMC3932189. Pubmed.
18. Taylor T; Tai CH; Huang J; Block J; Bai H; Kryshtafovych A; Montelione GT; Lee B. PROTEINS: Struc Funct Genomics. 2014, 82: Suppl 2: 14 – 25. Definition and classification of evaluation units for CASP10. PMC4133092. Pubmed.
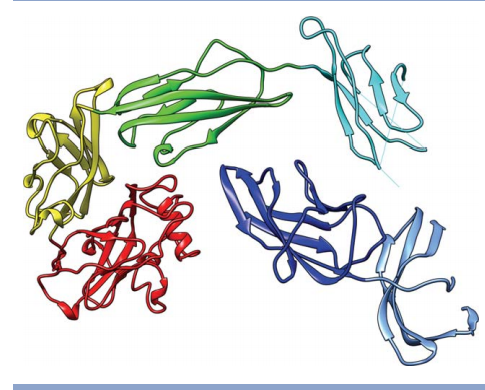
19. Snyder DA; Grullon J; Huang Y.J; Tejero R; Montelione GT. PROTEINS: Struc Funct Genomics. 2014, 82: Suppl 2: 219 – 230. The expanded FindCore method for identification of a core atom set for assessment of protein structure prediction. suppl. material. PMC3932188. Pubmed.
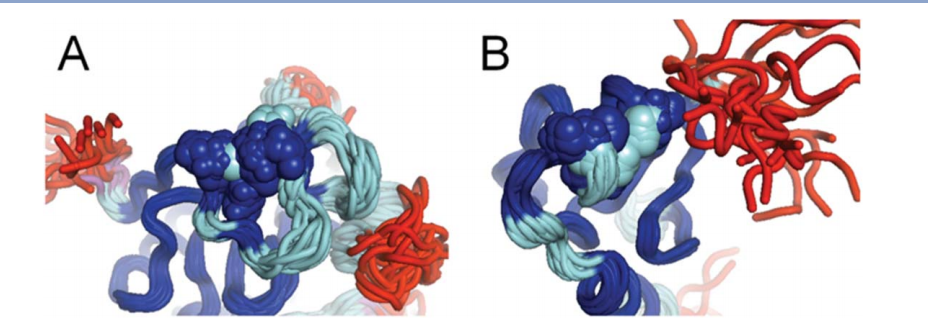
2013
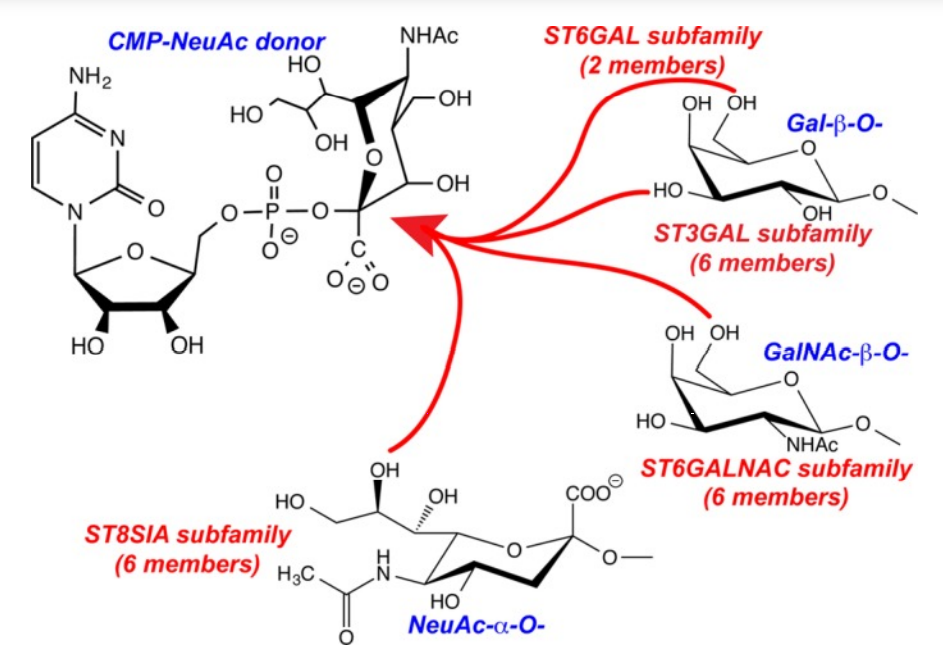
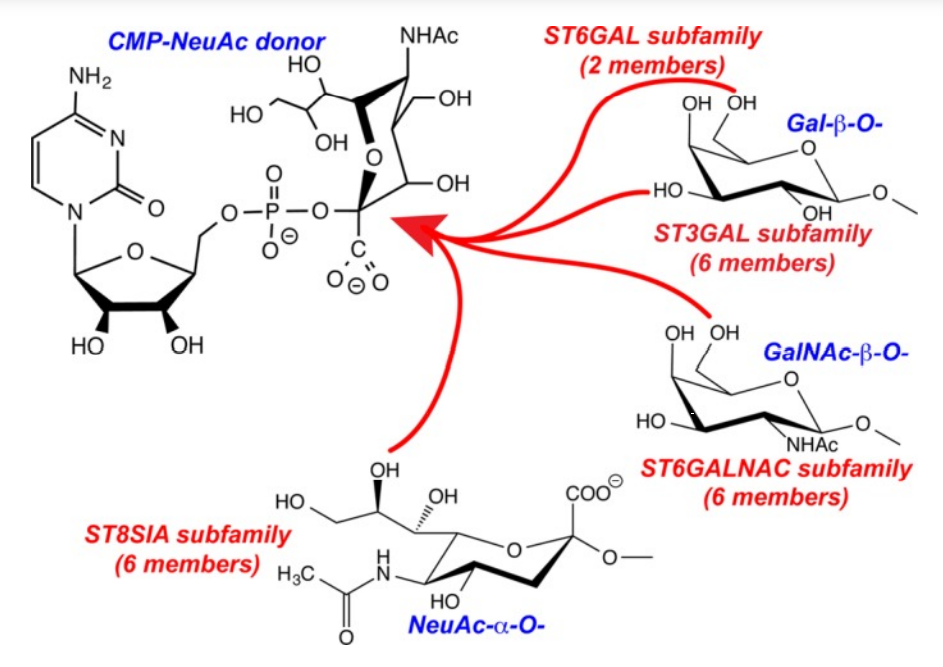
1. Meng L; Forouhar F; Thieker D; Gao Z; Ramiah A; Moniz H; Seetharaman J; Milaninia S; Su M; Bridger R; Veillon L; Azadi P; Kornhaber G; Wells L; Montelione G; Woods RJ; Tong L; Moremen KW. J Biol Chem. 2013, 288: 34680 – 34698. Enzymatic basis for N-glycan sialylation: structure of ST6GAL1 reveals conserved and unique features for glycan sialylation. suppl. material. PMC3843080. Pubmed.
2. Pulavarti S; He Y; Feldmann EA; Eletsky A; Acton TB; Xiao R; Everett JK; Montelione GT; Kennedy MA; Szyperski T. J Struct Funct Genomics. 2013, 14: 119 – 126. Solution NMR structures provide first structural coverage of the large protein domain family PF08369 and complementary structural coverage of dark operative protochlorophyllide oxidoreductase complexes. suppl. material. PMC3982801. Pubmed.
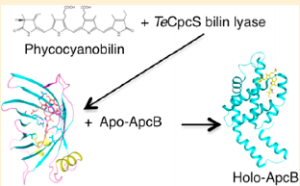
3. Kronfel CM; Kuzin AP; Forouhar F; Biswas A; Su M; Lew S; Seetharaman J; Xiao R; Everett JK; Ma LC; Acton TB; Montelione GT; Hunt JF; Paul CEC; Dragomani TM; Boutaghou, MN; Cole RB; Riml C; Alvey RM; Bryant DA; Schluchter WM. Biochemistry. 2013, 52: 8663 – 8676. Structural and biochemical characterization of the bilin lyase CpcS from Thermosynechococcus elongates. suppl. material. PMC3932240. Pubmed.
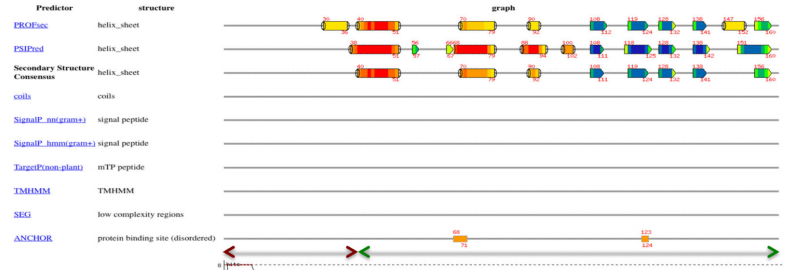
4. Huang Y.J; Acton TB; Montelione GT. Methods in Mol Biol. 2013, 1091: 3 – 16. DisMeta – a meta server for construct design and optimization. PMC4115584. Pubmed.

5. Uemura Y; Nakagawa N; WakamatsuT; Kim K; Montelione GT; Hunt JF; Kuramitsu S; Masui R. FEBS Lett. 2013, 587: 2669 – 2674. Crystal structure of the ligand-binding form of nanoRNase from Bacteroides fragilis, a member of the DHH/DHHA1 phosphoesterase family of proteins. suppl. material. PMC4113422. Pubmed.
6. Rossi P; Barbieri CM; Aramini JM; Bini E; Xiao R; Acton TB; Montelione GT. Nucleic Acids Research. 2013, 41: 2756 – 2768. Structures of apo- and ssDNA-bound YdbC from Lactococcus lactis uncover the role of protein family DUF2128 and expand the single-stranded DNA binding domain proteome. suppl. material. PMC3575825. Pubmed.
7. Mills J; Acton TB; Xiao R; Everett JK; Montelione GT; Szyperski T. J Struct Funct Genomics. 2013, 14: 19 – 24. Solution NMR structure of the helicase associated domain BVU_0683(627-691) from Bacteroides vulgatus provides first structural coverage for protein domain family PF03457 and indicates domain binding to DNA. suppl. material. PMC3637686. Pubmed.
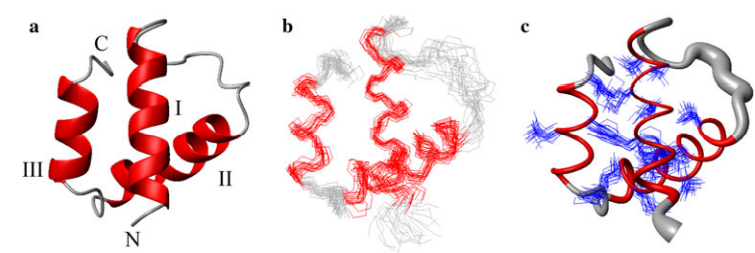
8. Bjelic S; Nivón LG; Celebi-Ölçüm N; Kiss G; Rosewall CF; Lovick HM; Ingalls EL; Gallaher JL; Seetharaman J; Lew S; Montelione GT; Hunt J.F.; Michael, F.E.; Houk, K.N.; Baker, D. ACS Chem. Biol. 2013, 8: 749 – 757. Computational design of enone-binding proteins with catalytic activity for the Morita-Baylis-Hillman reaction. suppl. material. PMC3647451. Pubmed.
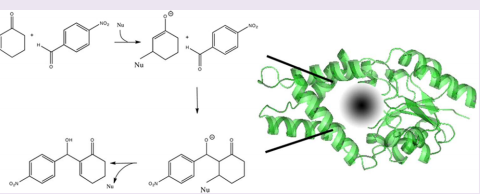
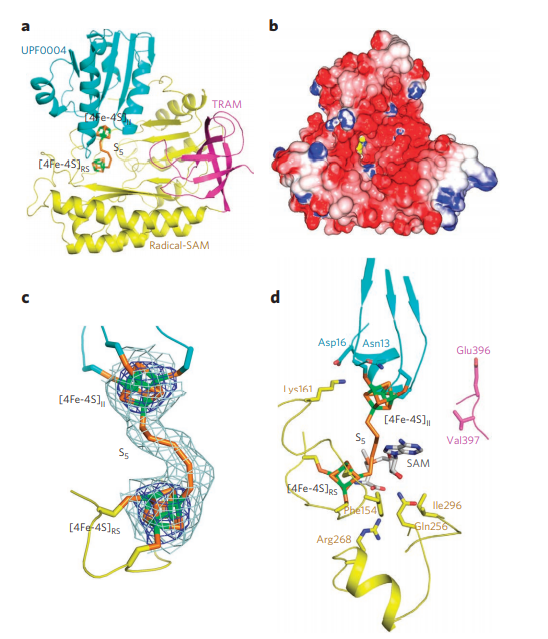
9. Forouhar F; Arragain S; Atta M; Gambarelli S; Mouesca JM; Hussain M; Xiao R; Kieffer-Jaquinod S; Seetharaman J; Acton TB; Montelione GT; Mulliez E; Hunt JF; Fontecave M. Nature Chemical Biology. 2013, 9: 333 – 338. Two Fe-S clusters catalyze sulfur insertion by radical-SAM methylthiotransferases. suppl. material. PMC4118475. Pubmed.
10. Froese DS; Forouhar F; Tran TH; Vollmar M; Kim Y; Lew S; Neely H; Seetharaman J; Shen Y; Xiao R; Acton TB; Everett JK; Cannone G; Puranik S; Savitsky P; Krojer T; Pilka ES; Kiyani W; Lee WH; Marsden BD; von Delft F; Allerston CK; Spagnolo L; Gileadi O; Montelione GT; Oppermann U; Yue WW; Tong L. Structure (Cell Press). 2013, 21: 1182 – 1192. Crystal structures of malonyl-CoA decarboxylase provide insights into its catalytic mechanism and disease-causing mutations. suppl. material. PMC3701320. Pubmed.

11. Procko E; Hedman R; Hamilton K; Seetharaman J; Fleishman S; Su M; Aramini J; Kornhaber G; Hunt J; Tong L; Montelione G; Baker D. J Mol Biol. 2013, 425: 3563 – 3575. Computational design of a protein-based enzyme inhibitor. suppl. material. PMC3818146. Pubmed.
12. Tejero R; Snyder D; Mao B; Aramini JM; Montelione GT; J Biomol NMR. 2013, 56: 337 – 351. PDBStat: A universal restraint converter and restraint analysis software package for protein NMR. suppl. material. PMC3932191. Pubmed.
13. Rosato A; Tejero R; Montelione GT. Current Opinions in Structural Biology. 2013, 23: 715 – 724. Quality assessment of protein NMR structures. PMC4110634. Pubmed.
14. Montelione GT; Nilges M; Bax A; Güntert P; Herrmann T; Richardso JS; Schwieters C; Vranken WF; Vuister GW; Wishart DS; Berman H;. Kleywegt GJ; Markley JL. Structure (Cell Press). 2013, 21: 1563 – 1570. Recommendations of the wwPDB NMR validation task force. PMC3884077. Pubmed.
15. Neklesa TK; Noblin DJ; Kuzin AP; Lew S; Seetharaman J; Acton TB; Kornhaber G; Xiao R; Montelione GT; Tong L; Crews CM. ACS Chem Biol. 2013, 8: 2293 – 2300. A bidirectional system for the dynamic small molecule control of intracellular fusion proteins. suppl. material. PMC4113957. Pubmed.
16. Pulavarti S; Eletsky A; Lee HW; Acton TB; Xiao R; Everett JK; Prestegard JH; Montelione GT; Szyperski T. J Struct Funct Genomics. 2013, 14: 155 – 160. Solution NMR structure of CD1104B from pathogenic Clostridium difficile reveals a distinct αhelical architecture and provides first structural representative of protein domain family PF14203. suppl. material. PMC3844015. Pubmed.
17. Ramelot TA; Yang Y; Sahu I.D; Lee HW; Xiao R; Lorigan GA; Montelione GT; Kennedey M. FEBS Lett. 2013, 587: 3522 – 3528. NMR structure and MD simulations of the AAA protease intermembrane space domain indicates peripheral membrane localization within the hexaoligomer. suppl. material. PMC4043124.
![(E and F) ConSurf [17] image showing the conserved surface residues. Residue coloring reflects the degree of residue conservation for selected eukaryotic homologs from Pfam06480 (42 sequences).](https://montelionelab.chem.rpi.edu/wp-content/uploads/2020/12/image-38.png)
18. Xu SY; Kuzin AP; Seetharaman J; Gutjahr A; Chan SH; Chen Y; Xiao R; Acton TB; Montelione GT; Tong L. PLoS One. 2013, 8:e72114. Structure determination and biochemical characterization of a putative HNH endonuclease from Geobacter metallireducens GS-15. suppl. material. PMC3765158.
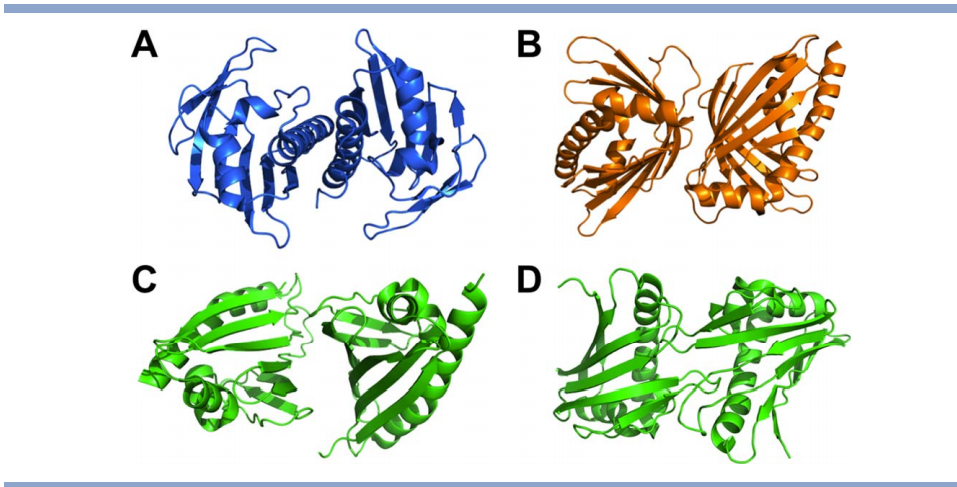
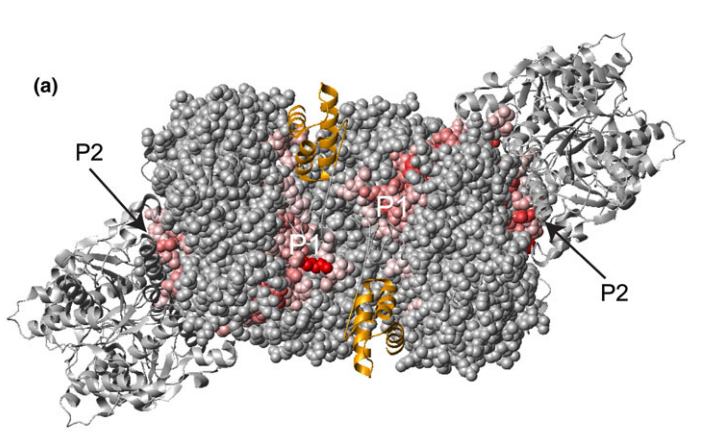
![Structure of Hpy99I dimer in complex with duplex DNA [18]. The catalytic domains are colored in cyan and green, and the N-terminal segment in yellow](https://montelionelab.chem.rpi.edu/wp-content/uploads/2020/12/image-39.png)
19. Meng L; Forouhar F; Thieker D; Gao Z; Ramiah A; Moniz H; Seetharaman J; Milaninia S; Su M; Bridger R; Veillon L; Azadi P; Kornhaber G; Wells L; Montelione G; Woods RJ; Tong L; Moremen KW. J Biol Chem. 2013, 288: 34680 – 34698. Enzymatic basis for N-glycan sialylation: structure of ST6GAL1 reveals conserved and unique features for glycan sialylation. suppl. material. PMC3843080. Pubmed.
20. Pulavarti S; He Y; Feldmann EA; Eletsky A; Acton TB; Xiao R; Everett JK; Montelione GT; Kennedy MA; Szyperski T. J Struct Funct Genomics. 2013, 14: 119 – 126. Solution NMR structures provide first structural coverage of the large protein domain family PF08369 and complementary structural coverage of dark operative protochlorophyllide oxidoreductase complexes. suppl. material. PMC3982801. Pubmed.
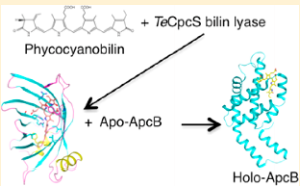
21. Kronfel CM; Kuzin AP; Forouhar F; Biswas A; Su M; Lew S; Seetharaman J; Xiao R; Everett JK; Ma LC; Acton TB; Montelione GT; Hunt JF; Paul CEC; Dragomani TM; Boutaghou, MN; Cole RB; Riml C; Alvey RM; Bryant DA; Schluchter WM. Biochemistry. 2013, 52: 8663 – 8676. Structural and biochemical characterization of the bilin lyase CpcS from Thermosynechococcus elongates. suppl. material. PMC3932240. Pubmed.
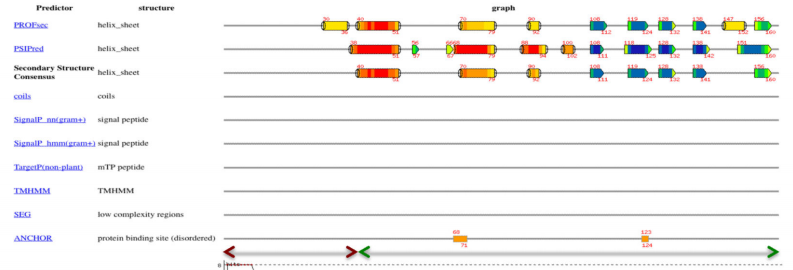
22. Huang Y.J; Acton TB; Montelione GT. Methods in Mol Biol. 2013, 1091: 3 – 16. DisMeta – a meta server for construct design and optimization. PMC4115584. Pubmed.
23. Uemura Y; Nakagawa N; WakamatsuT; Kim K; Montelione GT; Hunt JF; Kuramitsu S; Masui R. FEBS Lett. 2013, 587: 2669 – 2674. Crystal structure of the ligand-binding form of nanoRNase from Bacteroides fragilis, a member of the DHH/DHHA1 phosphoesterase family of proteins. suppl. material. PMC4113422. Pubmed.
2012
1. Thompson J; Sgourakis NG; Liu G; Rossi P; Tang Y; Mills J; Szyperski T; Montelione G; Baker D. Proc Natl Acad Sci USA. 2012, 109: 9875 – 9880. Accurate protein structure modeling using sparse NMR data and homologous structure information. suppl. material. PMC3382498. Pubmed.
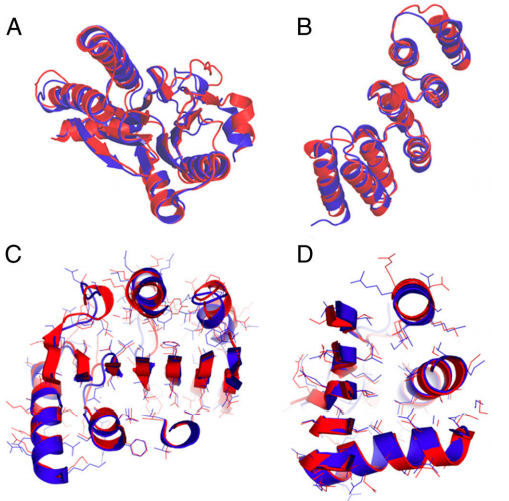
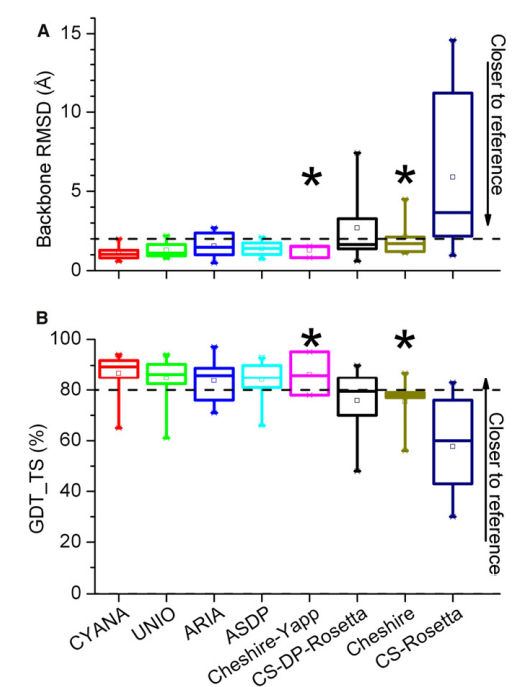
2. Rosato A; Aramini JM; Arrowsmith C; Bagaria A; Baker D; Cavalli A; Doreleijers JF; Eletsky A; Giachetti A; Guerry P; Gutmanas A; Güntert P; He Y; Herrmann T; Huang YJ; Jaravine V; Jonker HRA; Kennedy MA; Lange OF; Liu G; Malliavan TE; Mani R; Mao B; Montelione GT; Nilges M; Possi P; van der Schot G; Schwalbe H; Szyperski TA; Vendruscolo M; Vernon R; Vranken WF; de Vries S; Vuister GW; Wu B; Yang Y; Bonvin A.M.J.J. Structure (Cell Press) 2012, 20: 227 – 236. Blind testing of routine, fully automated determination of protein structures from NMR data. suppl. material 1. suppl. material 2. PMC3609704. Pubmed.
3. Ramelot TA; Yang Y; Xiao R; Acton TB; Everett JK; Montelione GT; Kennedy MA. PROTEINS: Struct Funct Bioinformatics. 2012, 2: 667 – 670. Solution NMR structure of BT_0084, a conjugative transposon lipoprotein from Bacteroides thetaiotamicron. suppl. material. PMC3766420. Pubmed.
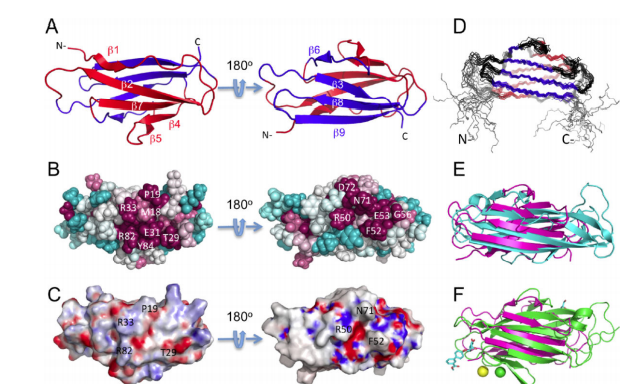
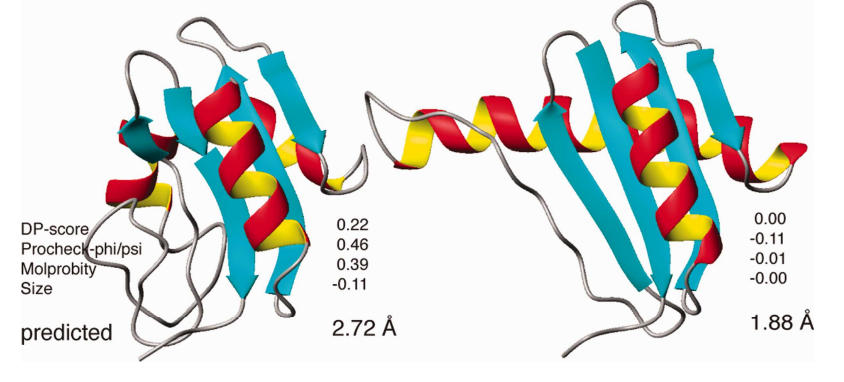
4. Bagaria A; Jaravine V; Huang YJ; Montelione GT; Güntert P. Protein Science. 2012, 21: 229 – 238. Protein structure validation by generalized linear model root-mean-square deviation prediction. suppl. material. PMC3324767. Pubmed.
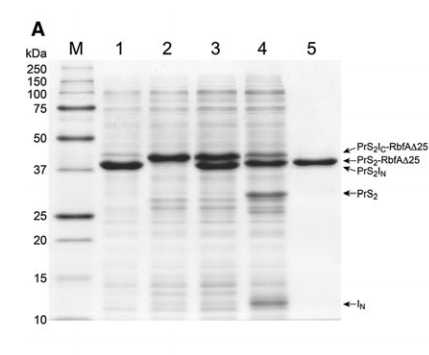
5. Kobayashi H; Swapna GVT; Wu KP; Afinogenova Y; Conover K; Mao B; Montelione GT; Inouye M. J Biomol NMR. 2012, 52: 303 – 313. Segmental isotope labeling of proteins for NMR structural study using a protein S tag for higher expression and solubility. suppl. material 1. suppl. material 2. PMC4117381. Pubmed.
6. Eletsky A; Petrey D; Zhang QC; Lee HW; Acton T; Xiao R; Everett J; Prestegard J; Honig B; Montelione GT; Szyperski T. J Struct Funct Genomics. 2012, 13: 1 – Solution NMR structures reveal unique homodimer formation by a winged helix-turn-helix motif and provide first structures for protein domain family PF10771. suppl. material. PMC3654790. Pubmed.
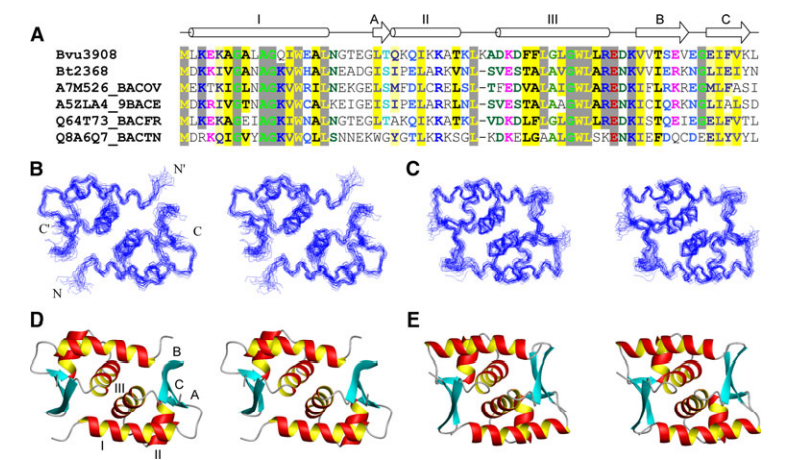
7. Eletsky A; Acton T; Xiao R; Everett J; Montelione GT; Szyperski T. J Struct Funct Genomics. 2012, 13: 9 – Solution NMR structures reveal a distinct architecture and provide first structures for protein domain family PF04536. suppl. material. PMC360422. Pubmed.
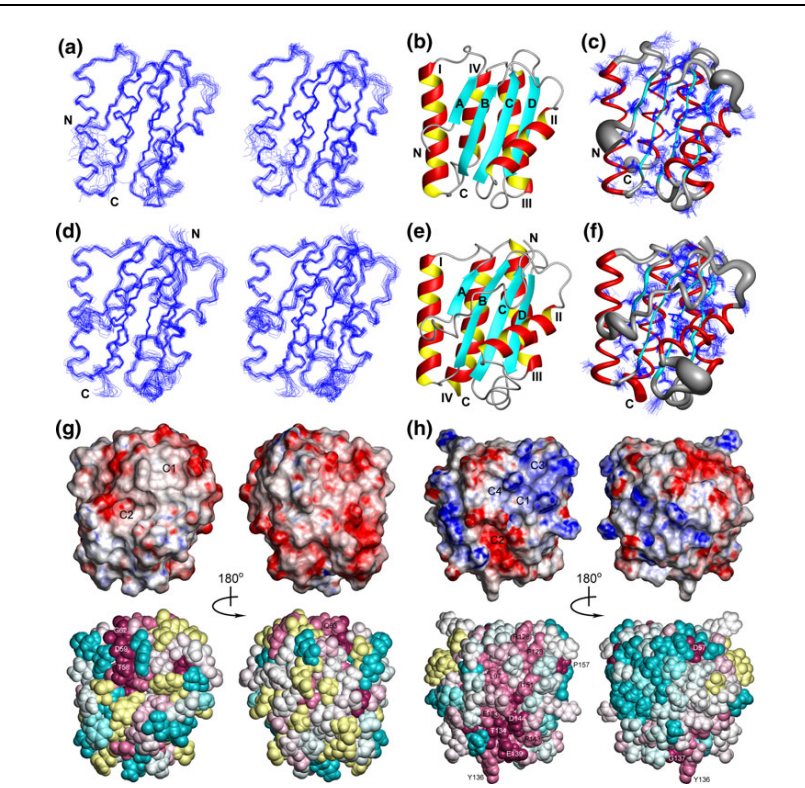
8. Wu Y; Punta M; Xiao R; Acton T; Sathyamoorthy B; Dey F; Fischer M; Skerra A; Rost B; Montelione GT; Szyperski T. PLOS One. 2012, 7: e37404. NMR structure of lipoprotein YxeF from Bacillus subtilis reveals a calycin fold and distant homology with the lipocalin Blc from Escherichia coli. PMC3367933. Pubmed.
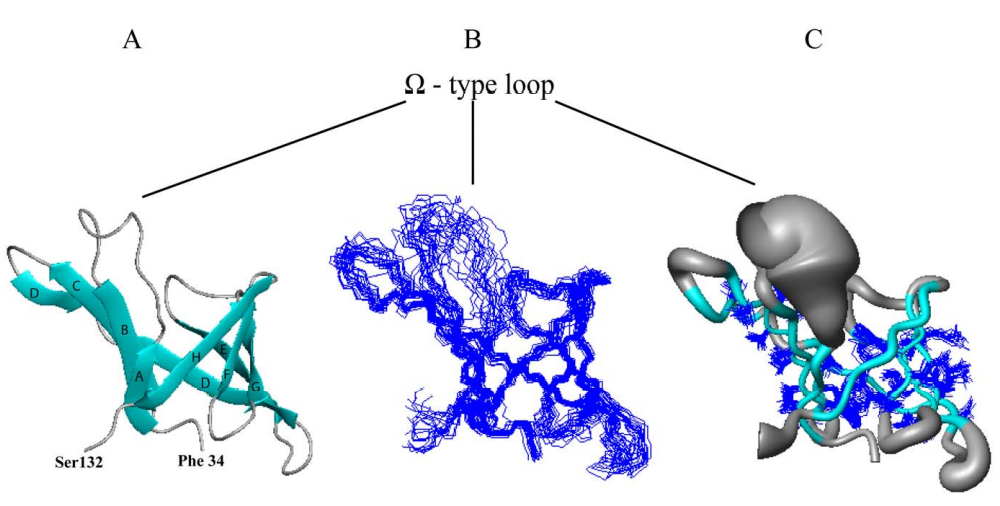
9. Ertekin A; Aramini JM; Rossi P; Leonard PG; Janjua H; Xiao R; Maglaqui M; Lee HW; Prestegard JH; Montelione GT. J Biol Chem. 2012, 287: 16541 – 16549. Human cyclin dependent kinase 2 associated protein 1 is dimeric in its disulfide-reduced state, with natively disordered n-terminal region. suppl. material. PMC3351331. Pubmed.
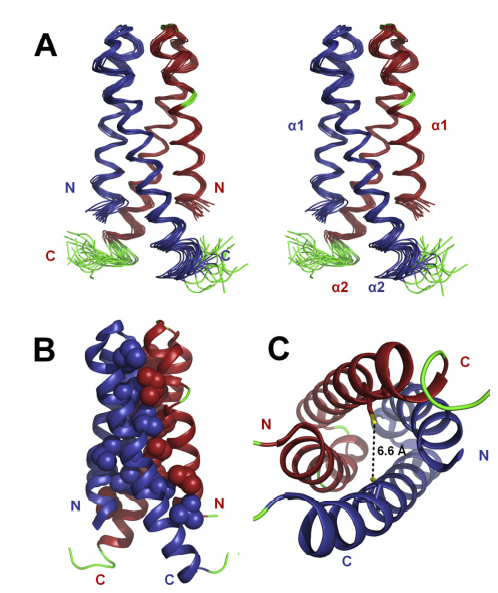
10. Huang Y; Rosato A; Singh G; Montelione GT. Nucleic Acids Research. 2012, 40: W542 – 546. RPF – A quality assessment tool for protein NMR structures. PMC3394279. Pubmed.
11. Snyder D; Aramini JM; Yu B; Huang Y; Xiao R; Cort J; Shastry R; Ma LM; Liu J; Rost B; Action T; Kennedy M; Montelione GT. PROTEINS: Struct Funct Genomics. 2012, 80: 1901 – 1906. Solution NMR structure of the ribosomal protein RP-L35Ae from Pyrococcus furiosus. suppl. material. PMC3639469. Pubmed.
12. Montelione GT. Faculty 1000 Commentary. 2012, 4: 7. The Protein Structure Initiative: achievements and visions for the future. PMC3318194.
13. Lange OF; Rossi P; Sgourakis N; Song Y; Lee HW; Aramini JM; Ertekin A; Xiao R; Acton TB; Montelione GT; Baker D. Proc Natl Acad Sci U.S.A. 2012, 109: 10873 – 10878. The determination of solution structures of proteins up to 40 kDa using CS- Rosetta with sparse NMR data from deuterated samples. PMC3390869.
14. Feldmann EA; Seetharaman J; Ramelot TA; Lew S; Zhao L; Hamilton K; Ciccosanti C; Xiao R; Acton TB; Everett JK; Tong L; Montelione GT; Kennedy MA. J Struct Funct Genomics. 2012, 13: 155 – 162. Solution NMR and X-ray crystal structure of Pspto_3016 from Pseudomonas syringae, a member of protein domain family PF04237 (DUF419) that adopts a “double wing” DNA binding motif. suppl. material. PMC3697073.
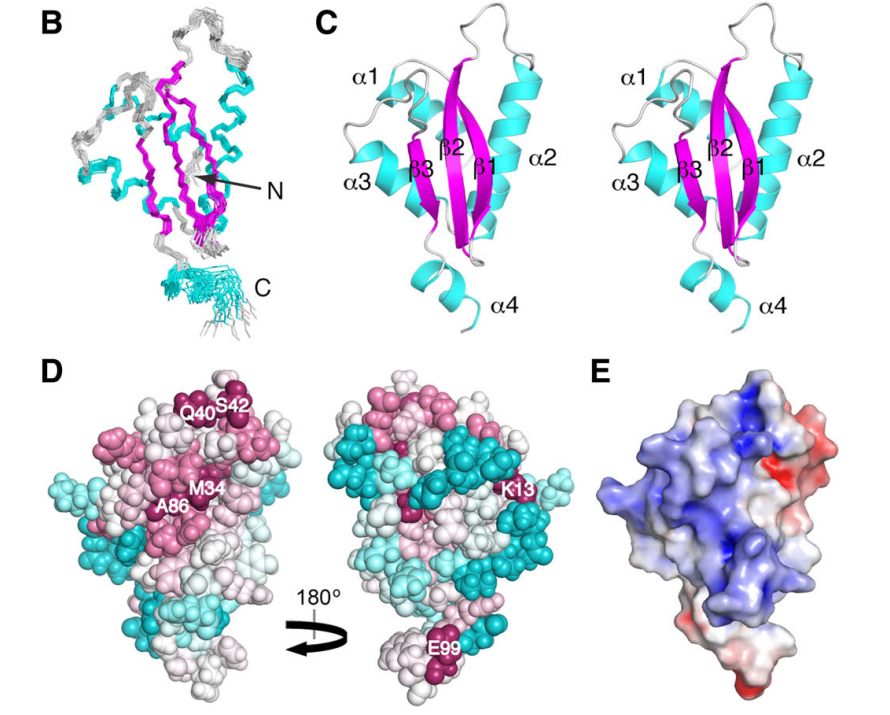
15. Vorobiev SM; Neely H; Yu B; Seetharaman J; Xiao R; Acton TB; Montelione GT; Hunt JF. J Struct Funct Genomics. 2012, 13: 177 – 183. Crystal structure of a catalytically active GG(D/E)EF diguanylate cyclase domain from Marinobacter aquaeolei with bound c-di-GMP product. suppl. material. PMC3683829. Pubmed.
16. Swapna GVT; Rossi P; Montelione AF; Benach J; Yu B; Abashidze M; Seetharaman J; Xiao R; Acton TB; Tong L; Montelione GT. J Struct Funct Genomics. 2012, 13: 163 – 170. Three structural representatives of the PF06855 protein domain family from Staphyloccocus aureus and Bacillus subtilis have SAM domain- like folds and different functions. PMC4075964. Pubmed.
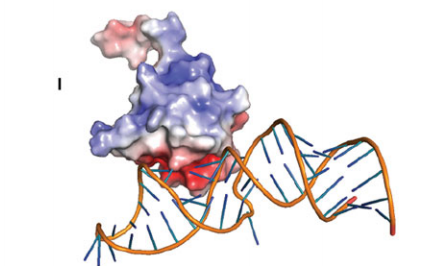
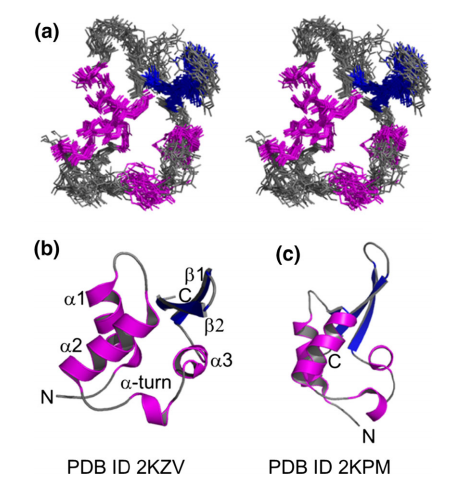
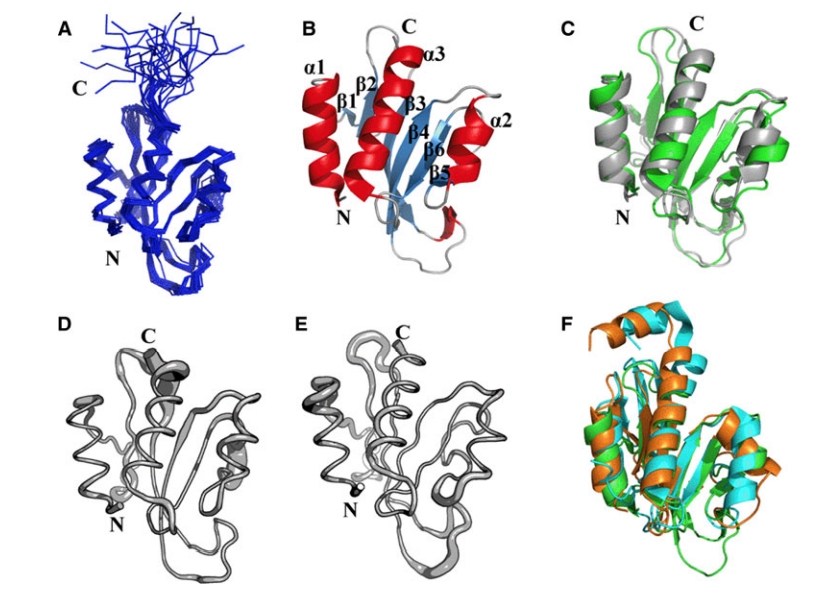
17. Aramini JM; Petrey D; Lee DY; Janjua H; Xiao R; Acton TB; Everett JK; Montelione G.T. J Struct Funct Genomics. 2012, 13: 171 – 176. Solution NMR structure of Alr2454 from Nostoc sp. PCC 7120, the first structural representative of Pfam domain family PF11267. suppl. material. PMC3897273. Pubmed.
![image showing the conserved residues in Alr2454 (residues 3–101). Residue coloring, reflecting the degree of residue conservation over the entire PF11267 protein domain family (Pfam 25.0 [1]; 80 sequences), ranges from magenta (highly conserved) to cyan (variable).](https://montelionelab.chem.rpi.edu/wp-content/uploads/2020/12/image-61-1024x471.png)
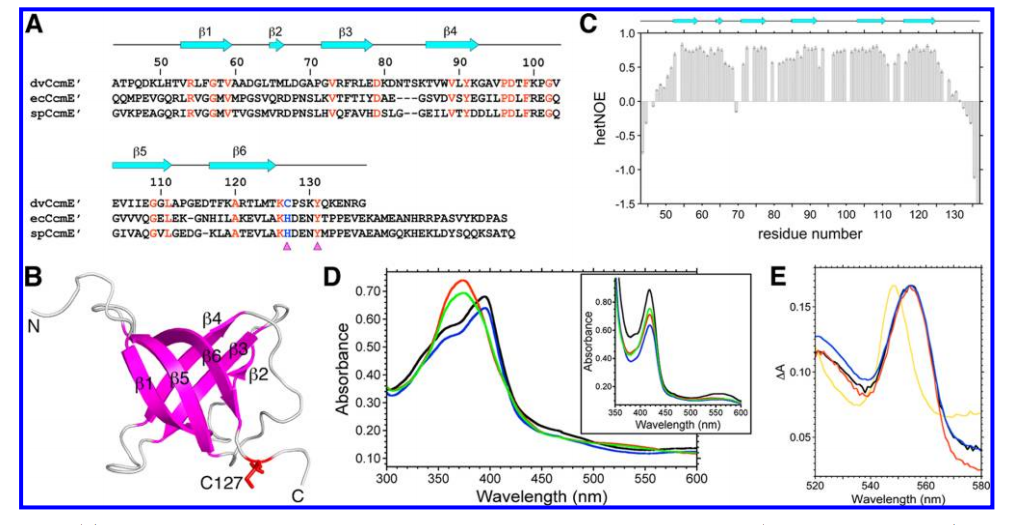
18. Aramini JM; Hamilton K; Rossi P; Ertekin A; Lee HW; Lemak A; Wang H; Xiao R; Acton TB; Everett JK; Montelione GT. Biochemistry. 2012, 51: 3705 – 3707. Solution NMR structure, backbone dynamics, and heme-binding properties of a novel cytochrome c maturation protein CcmE from Desulfovibrio vulgaris. suppl. material. PMC3366507. Pubmed.
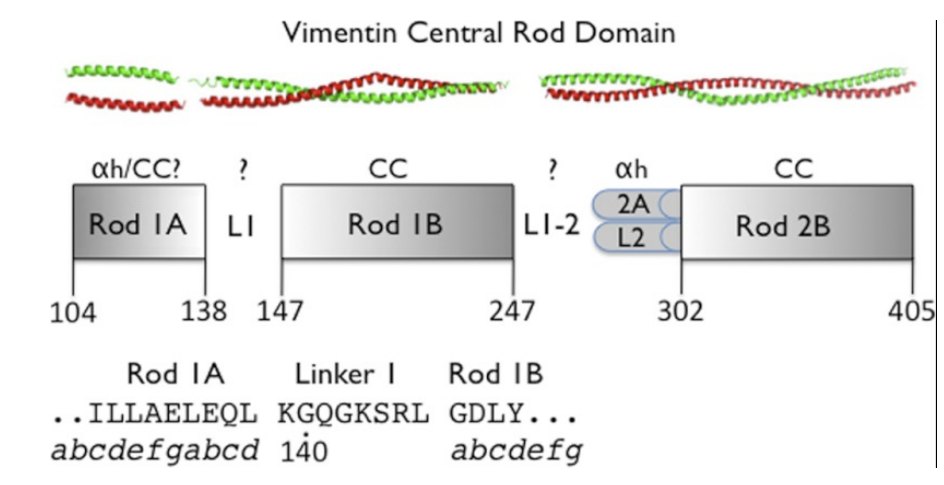
19. Aziz A; Hess JF; Budamagunta MS; Voss JC; Kuzin AP; Huang YJ; Xiao R; Montelione GT; Fitzgerald PG; Hunt JF. J Biol Chem. 2012, 287: 28349 – 283461. The structure of vimentin linker 1 and rod 1B domains characterized by site-directed spin-labeling electron paramagnetic resonance (SDSL-EPR) and x-ray crystallography. PMC3436525. Pubmed.
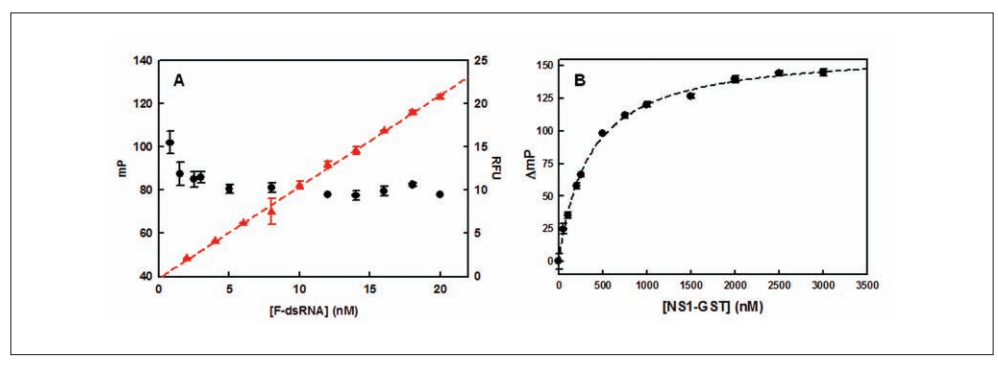
20. Cho EJ; Xia S; Ma LC; Robertus J; Krug RM; Anslyn EV; Montelione GT; Ellington AD. J Biomol Screen. 2012, 17: 448 – 459. Identification of influenza virus inhibitors targeting NS1A utilizing fluorescence polarization-based high-throughput assay. Pubmed.

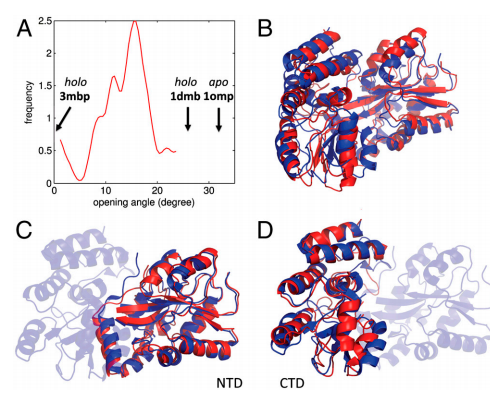
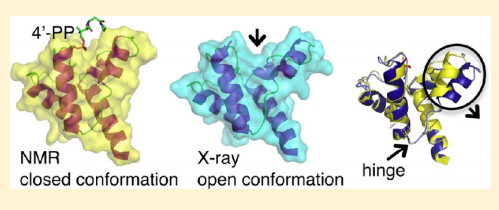
21. Ramelot TA; Rossi P; Forouhar F; Lee HW; Yang Y; Ni S; Unser S; Lew S; Seetharaman J; Xiao R; Acton TB; Everett JK; Prestegard JH; Hunt JF; Montelione GT; Kennedy MA. Biochemistry. 2012, 51: 7239 – 7249. Structure of a specialized acyl carrier protein essential for lipid A biosynthesis with very long chain fatty acids in open and closed conformations. suppl. material. PMC4104962. Pubmed.
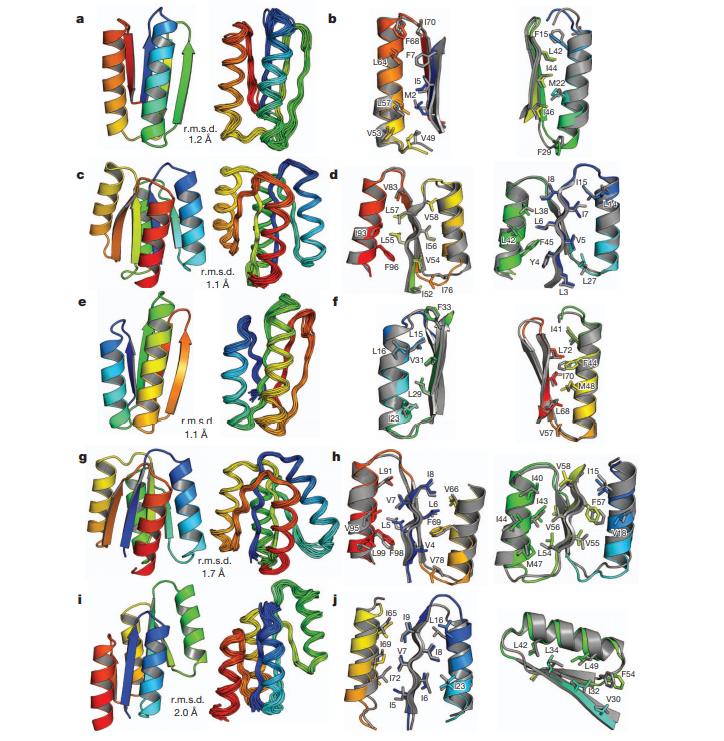
22. Koga N; Tatsumi-Koga R; Liu G; Xiao R; Acton TB; Montelione GT; Baker D. Nature. 2012, 491: 222 – 227. Principles for designing ideal protein structures. supplmaterial. PMC3705962. Pubmed.
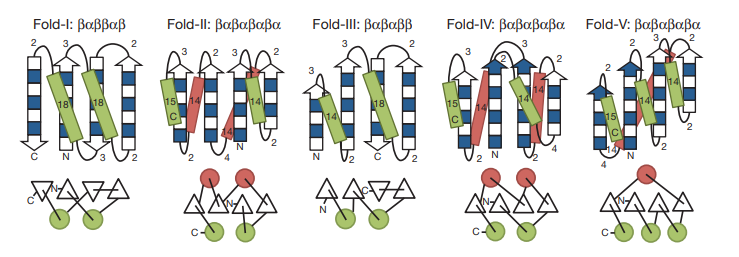
23. Montelione GT. Faculty 1000 Commentary. 2012, 4: 7. The Protein Structure Initiative: achievements and visions for the future. PMC3318194.
24. Lange OF; Rossi P; Sgourakis N; Song Y; Lee HW; Aramini JM; Ertekin A; Xiao R; Acton TB; Montelione GT; Baker D. Proc Natl Acad Sci USA. 2012, 109: 10873 – 10878. The determination of solution structures of proteins up to 40 kDa using CS- Rosetta with sparse NMR data from deuterated samples. suppl. material. PMC3390869. Pubmed.
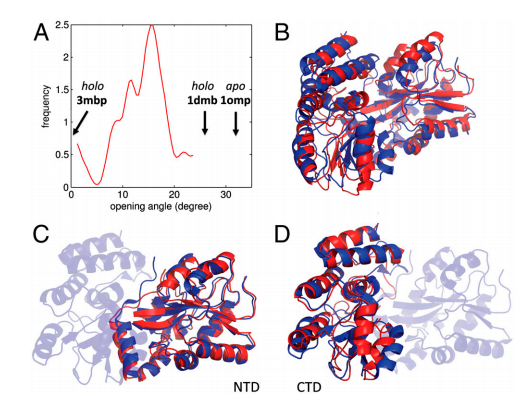
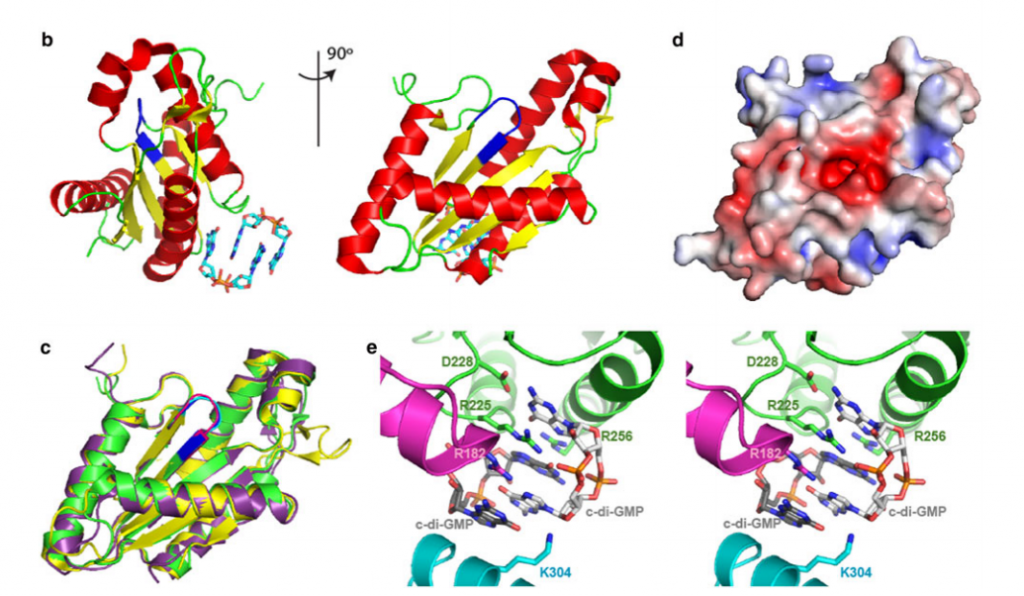
25. Vorobiev SM; Neely H; Yu B; Seetharaman J; Xiao R; Acton TB; Montelione GT; Hunt J. J Struct Funct Genomics. 2012, 13: 177 – 183. Crystal structure of a catalytically active GG(D/E)EF diguanylate cyclase domain from Marinobacter aquaeolei with bound c-di-GMP product. suppl. material. PMC3683829. Pubmed.
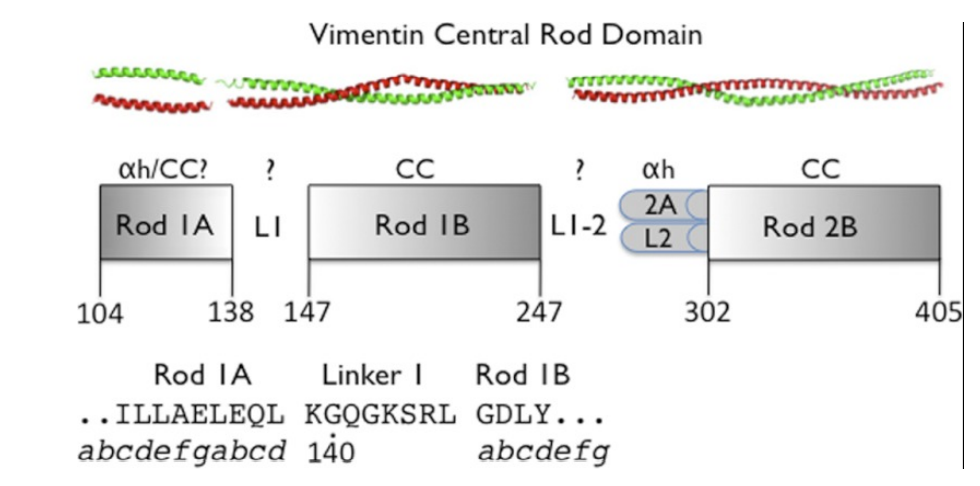
26. Aziz A; Hess JF; Budamagunta MS; Voss JC; Kuzin AP; Huang Y.J; Xiao R; Montelione GT; Fitzgerald PG; Hunt JF. J Biol Chem. 2012, 287: 28349 – 283461. The structure of vimentin linker 1 and rod 1B domains characterized by site-directed spin-labeling electron paramagnetic resonance (SDSL-EPR) and x-ray crystallography. PMC3436525. Pubmed.
27. Cho E.J; Xia S; Ma LC; Robertus J; Krug RM; Anslyn EV; Montelione GT; Ellington AD. J Biomol Screen. 2012, 17: 448 – 459. Identification of influenza virus inhibitors targeting NS1A utilizing fluorescence polarization-based high-throughput assay. PMC in process. Pubmed.
28. Koga N; Tatsumi-Koga R; Liu G; Xiao R; Acton TB; Montelione GT; Baker D. Nature. 2012, 491: 222 – 227. Principles for designing ideal protein structures. suppl. material. PMC3705962. Pubmed.
29. Kim D; Zheng H; Huang YJ; Montelione GT; Hunt JF. J Amer Chem Soc. 2012, 135: 2999 – 3010. ATPase active-site electrostatic interactions control the global conformation of the 100 kDa SecA translocase. suppl. material. PMC4134686. Pubmed.
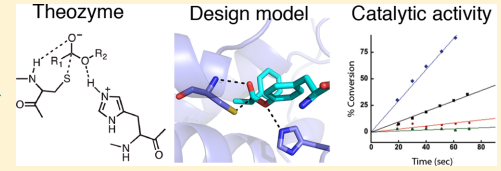
30. Richter F; Blomberg R; Khare SD; Kiss G; Kuzin AP; Smith AJ; Gallaher JL; Pianowski Z; Helgeson RC; Grjasnow A; Xiao R; Seetharaman J; Su M; Vorobiev S; Lew S; Forouhar F; Kornhaber GJ; Hunt JF; Montelione GT; Tong L; Houk KN; Hilvert D; Baker D. J Amer Chem Soc. 2012, 135: 16197 – 16206. Computational design of catalytic dyads and oxyanion holes for ester hydrolysis. suppl. material. PMC4104585. Pubmed.
31. Eletsky A; Jeong MY; Kim H; Lee HW; Xiao R; Pagliarini D.J; Prestegard JH; Winge DR; Montelione GT; Szyperski T. Biochemistry. 2012, 51: 8475 – 8477. Solution NMR structure of yeast succinate dehydrogenase flavinylation factor Sdh5 reveals a putative Sdh1 binding site. suppl. material. PMC3667956. Pubmed.
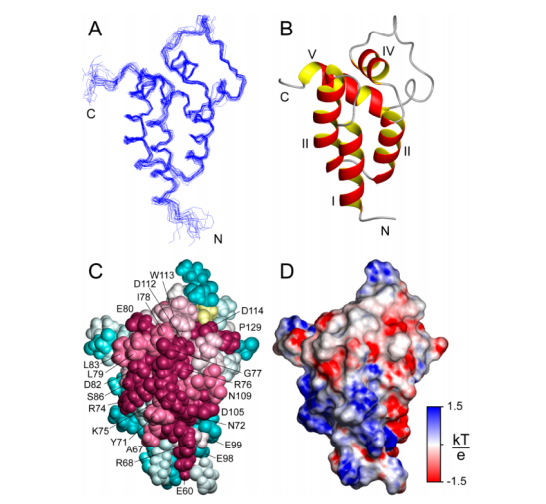
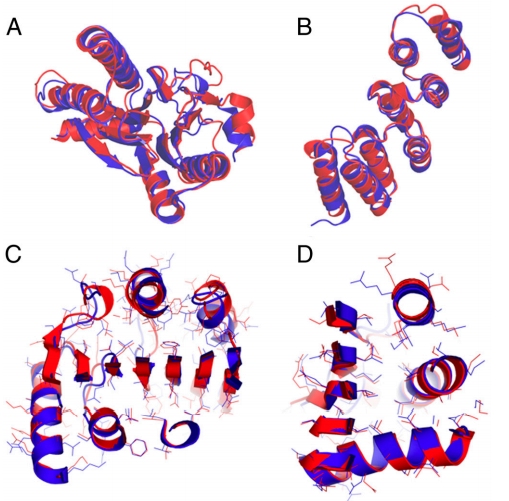
32. Thompson J; Sgourakis NG; Liu G; Rossi P; Tang Y; Mills J; Szyperski T; Montelione G; Baker D. Proc Natl Acad Sci USA. 2012, 109: 9875 – 9880. Accurate protein structure modeling using sparse NMR data and homologous structure information. suppl. material. PMC3382498. Pubmed.
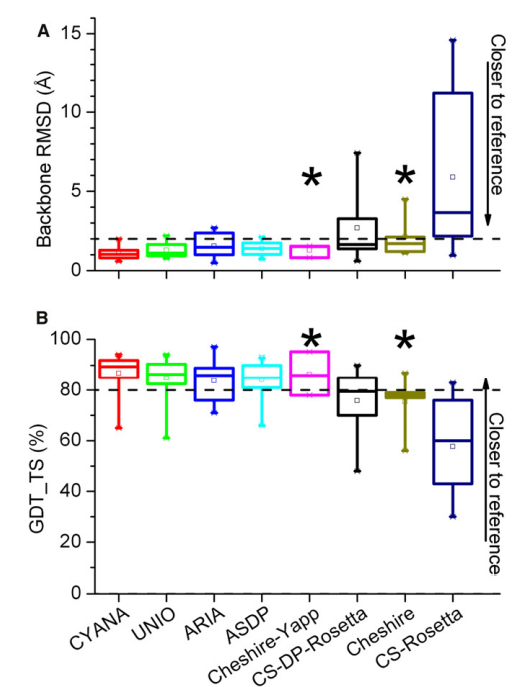
33. Rosato A; Aramini JM; Arrowsmith C; Bagaria A; Baker D; Cavalli A; Doreleijers JF; Eletsky A; Giachetti A; Guerry P; Gutmanas A; Güntert P; He Y; Herrmann T; Huang YJ; Jaravine V; Jonker HRA; Kennedy MA; Lange OF; Liu G; Malliavan TE; Mani R; Mao B; Montelione GT; Nilges M; Possi P; van der Schot G; Schwalbe H; Szyperski TA; Vendruscolo M; Vernon R; Vranken WF; de Vries S; Vuister GW; Wu B; Yang Y; Bonvin A.M.J.J. Structure (Cell Press) 2012, 20: 227 – 236. Blind testing of routine, fully automated determination of protein structures from NMR data. suppl. material 1. suppl. material 2. PMC3609704. Pubmed.
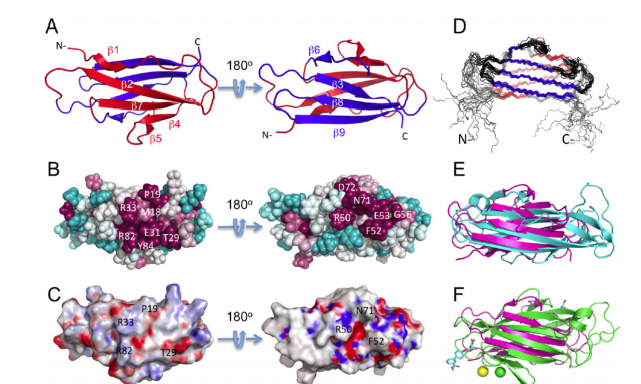
34. Ramelot TA; Yang Y; Xiao R; Acton TB; Everett JK; Montelione GT; Kennedy MA. PROTEINS: Struct Funct Bioinformatics. 2012, 2: 667 – 670. Solution NMR structure of BT_0084, a conjugative transposon lipoprotein from Bacteroides thetaiotamicron. suppl. material. PMC3766420. Pubmed.
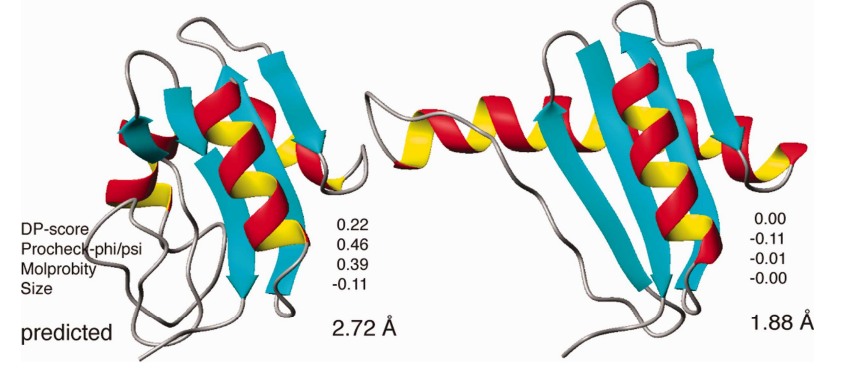
35. Bagaria A; Jaravine V; Huang YJ; Montelione GT; Güntert P. Protein Science. 2012, 21: 229 – 238. Protein structure validation by generalized linear model root-mean-square deviation prediction. suppl. material. PMC3324767. Pubmed.
36. Kobayashi H; Swapna GVT; Wu KP; Afinogenova Y; Conover K; Mao B; Montelione GT; Inouye M. J Biomol NMR. 2012, 52: 303 – 313. Segmental isotope labeling of proteins for NMR structural study using a protein S tag for higher expression and solubility. suppl. material 1. suppl. material 2. PMC4117381. Pubmed.
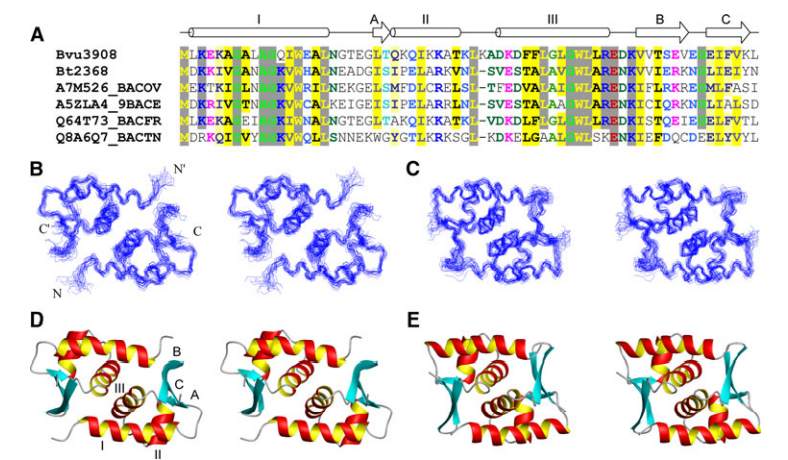
37. Eletsky A; Petrey D; Zhang QC; Lee HW; Acton T; Xiao R; Everett J; Prestegard J; Honig B; Montelione GT; Szyperski T. J Struct Funct Genomics. 2012, 13: 1 – Solution NMR structures reveal unique homodimer formation by a winged helix-turn-helix motif and provide first structures for protein domain family PF10771. suppl. material. PMC3654790. Pubmed.
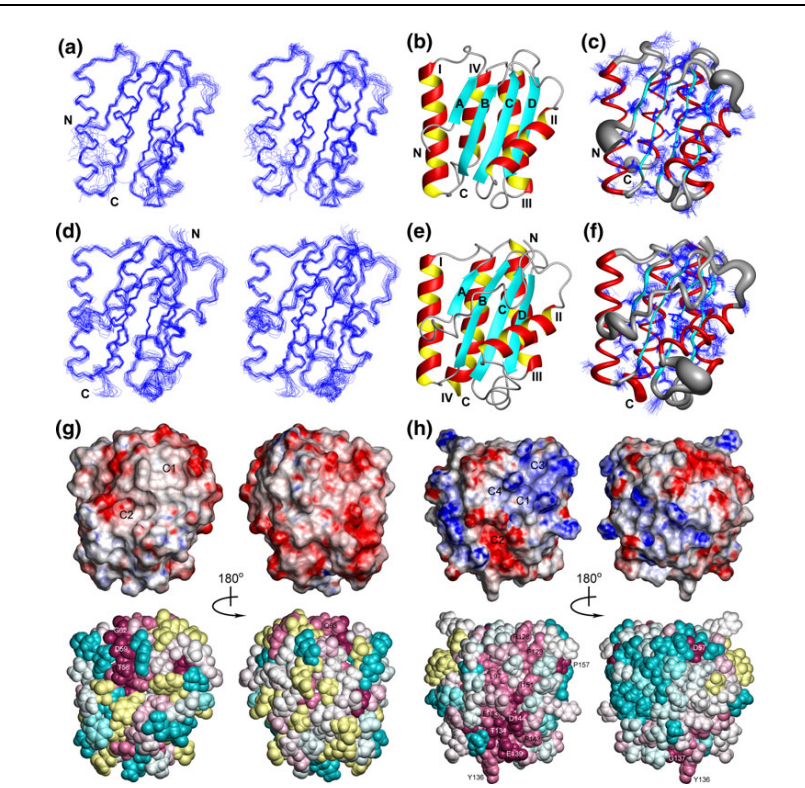
38. Eletsky A; Acton T; Xiao R; Everett J; Montelione GT; Szyperski T. J Struct Funct Genomics. 2012, 13: 9 – Solution NMR structures reveal a distinct architecture and provide first structures for protein domain family PF04536. suppl. material. PMC360422. Pubmed.
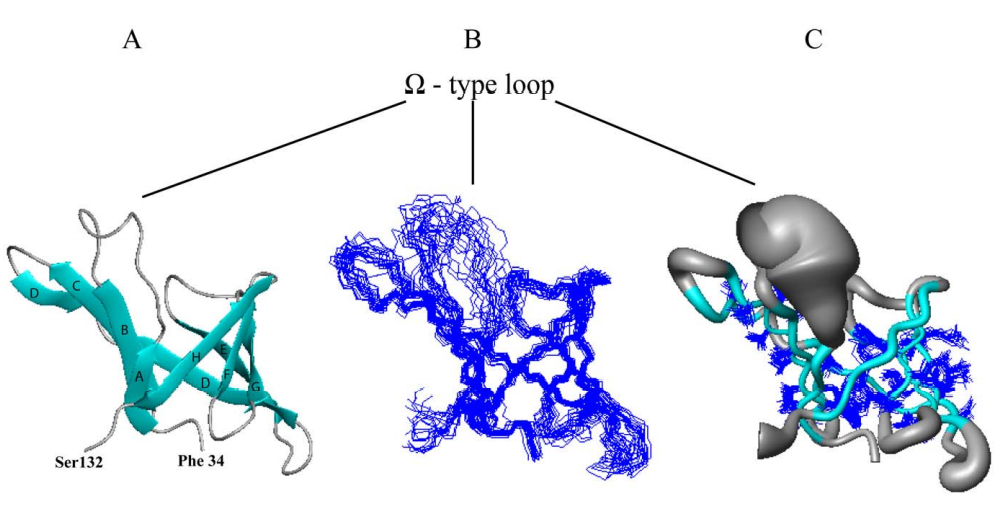
39. Wu Y; Punta M; Xiao R; Acton T; Sathyamoorthy B; Dey F; Fischer M; Skerra A; Rost B; Montelione GT; Szyperski T. PLOS One. 2012, 7: e37404. NMR structure of lipoprotein YxeF from Bacillus subtilis reveals a calycin fold and distant homology with the lipocalin Blc from Escherichia coli. PMC3367933. Pubmed.
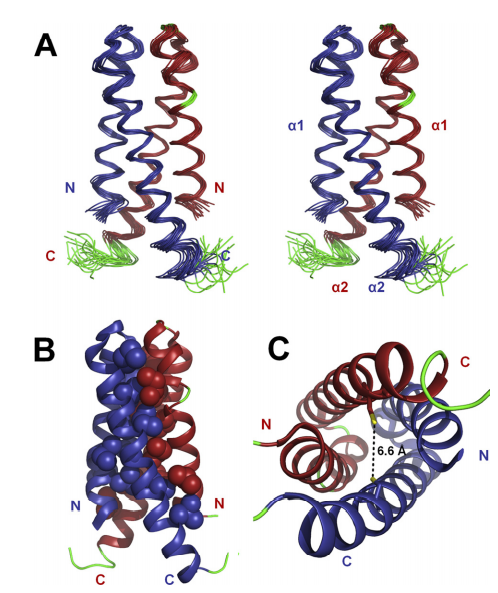
40. Ertekin A; Aramini JM; Rossi P; Leonard PG; Janjua H; Xiao R; Maglaqui M; Lee HW; Prestegard JH; Montelione GT. J Biol Chem. 2012, 287: 16541 – 16549. Human cyclin dependent kinase 2 associated protein 1 is dimeric in its disulfide-reduced state, with natively disordered n-terminal region. suppl. material. PMC3351331. Pubmed.
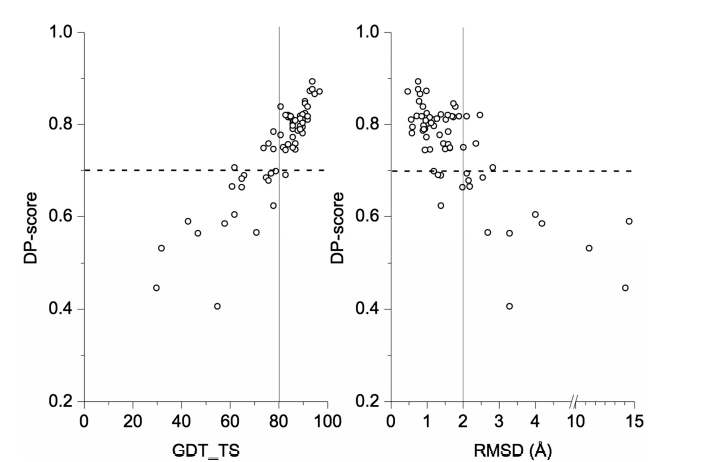
41. Huang Y; Rosato A; Singh G; Montelione GT. Nucleic Acids Research. 2012, 40: W542 – 546. RPF – A quality assessment tool for protein NMR structures. PMC3394279. Pubmed.
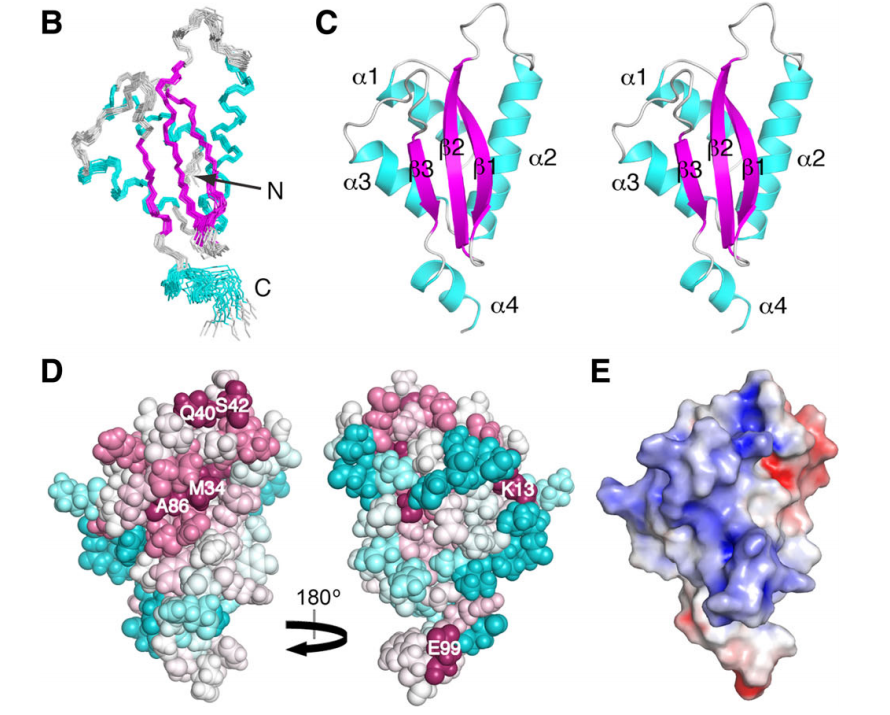
42. Snyder D; Aramini JM; Yu B; Huang Y; Xiao R; Cort J; Shastry R; Ma LM; Liu J; Rost B; Action T; Kennedy M; Montelione GT. PROTEINS: Struct Funct Genomics. 2012, 80: 1901 – 1906. Solution NMR structure of the ribosomal protein RP-L35Ae from Pyrococcus furiosus. suppl. material. PMC3639469. Pubmed.
43. Montelione GT. Faculty 1000 Commentary. 2012, 4: 7. The Protein Structure Initiative: achievements and visions for the future. PMC3318194.
44. Lange OF; Rossi P; Sgourakis N; Song Y; Lee HW; Aramini JM; Ertekin A; Xiao R; Acton TB; Montelione GT; Baker D. Proc Natl Acad Sci U.S.A. 2012, 109: 10873 – 10878. The determination of solution structures of proteins up to 40 kDa using CS- Rosetta with sparse NMR data from deuterated samples. PMC3390869.