2023
Kryshtafovych A, Montelione GT, Rigden DJ, Mesdaghi S, Karaca E, Moult J. Breaking the conformational ensemble barrier: Ensemble structure modeling challenges in CASP15. PROTEINS: Structure Function Bioinformatics Online. 1-9. doi:10.1002/prot.26584. 2023.
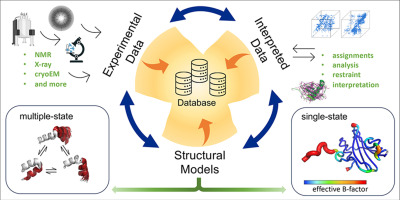
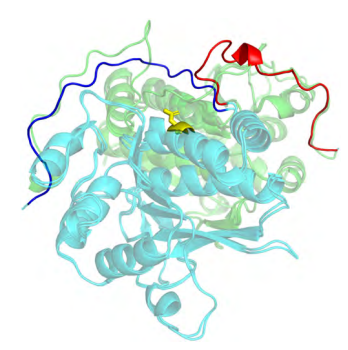
Ramelot TA, Tejero R, Montelione GT. Representing structures of the multiple conformational states of proteins. Curr. Opin. Struct. Biol. 83, 102703. 2023.
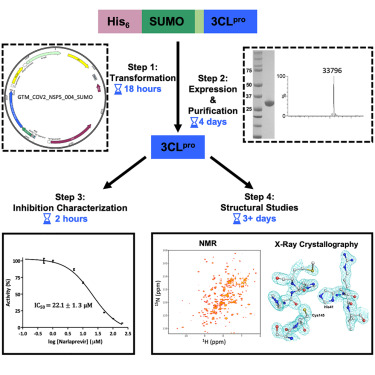
Mazzei L, Greene-Cramer R, Bafna K, Jovanovic A, De Falco A, Acton TB, Royer CA, Ciurli S, Montelione GT. Protocol for production and purification of SARS-CoV-2 3CLpro. STAR Protocols. 2023 May 5;4(2):102326-.
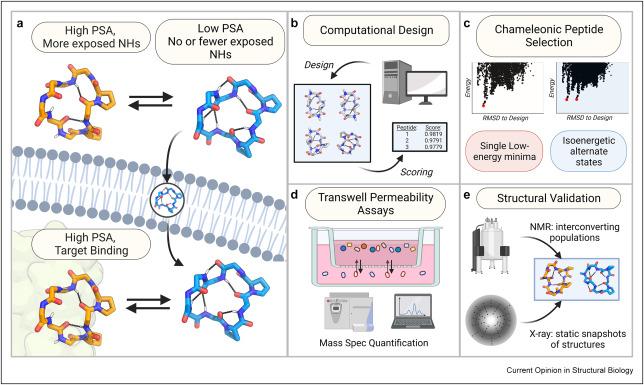
Ramelot TA, Palmer J, Montelione GT, Bhardwaj G. Cell-permeable chameleonic peptides: Exploiting conformational dynamics in de novo cyclic peptide design. Current Opinion in Structural Biology. 2023 Jun 1;80:102603.
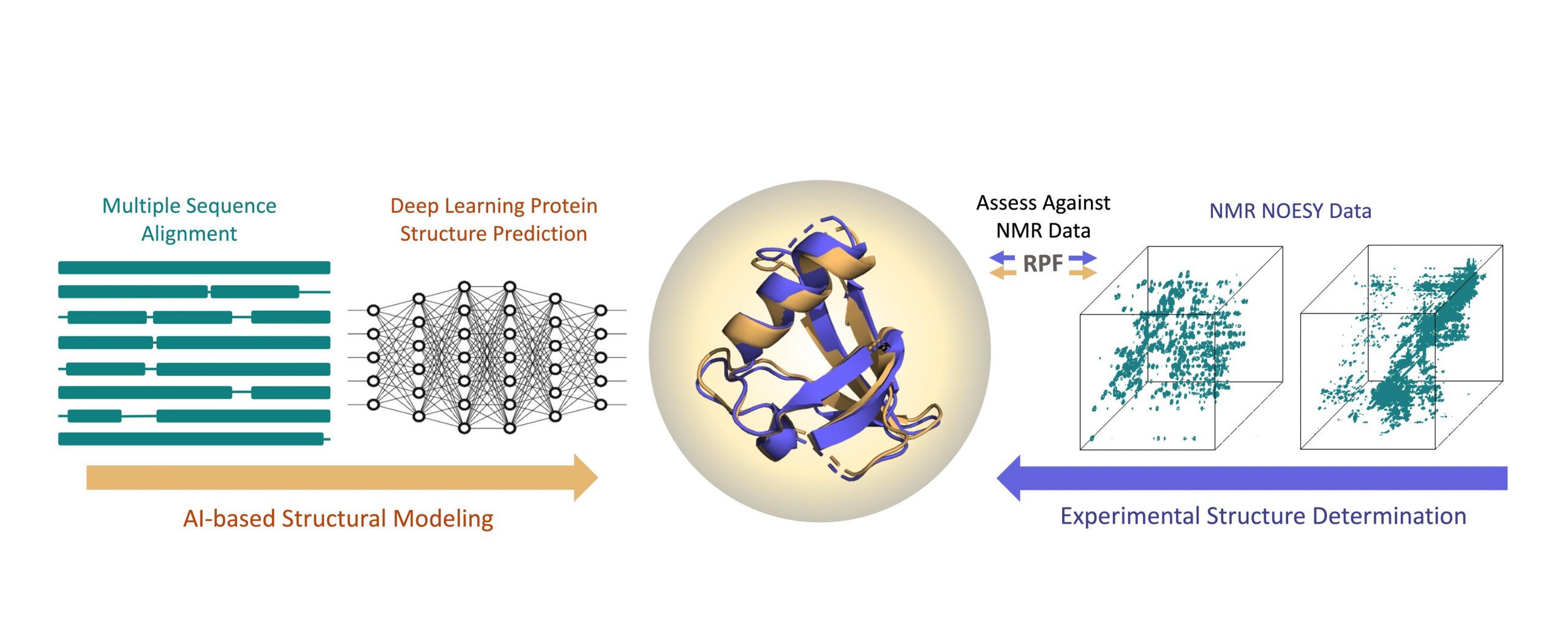
Li EH, Spaman LE, Tejero R, Huang YJ, Ramelot TA, Fraga KJ, Prestegard JH, Kennedy MA, Montelione GT. Blind assessment of monomeric AlphaFold2 protein structure models with experimental NMR data. Journal of Magnetic Resonance. 2023 May 20:107481.

Maisuradze GG, Montelione GT, Rackovsky S, Skolnick J. Protein Folding and Dynamics─ An Overview on the Occasion of Harold Scheraga’s 100th Birthday. The Journal of Physical chemistry. B. 2023 Apr 1;127(13):2879-80.
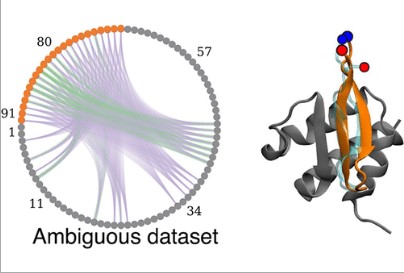
Mondal A, Swapna GV, Lopez MM, Klang L, Hao J, Ma L, Roth MJ, Montelione GT, Perez A. Structure Determination of Challenging Protein–Peptide Complexes Combining NMR Chemical Shift Data and Molecular Dynamics Simulations. ACS J. Chem Inform Modeling, 2023.. 2023 Mar 29;63(7):2058-72.
2022
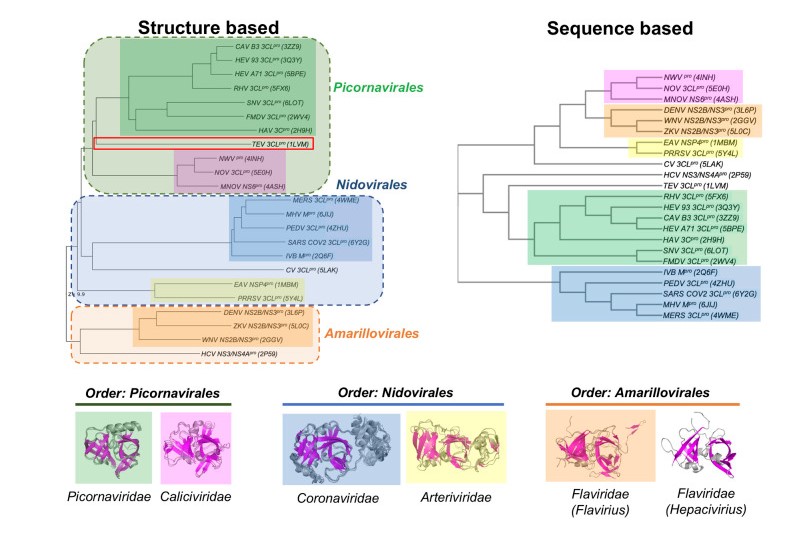
Khushboo Bafna, Christopher L Cioffi, Robert M Krug, Gaetano T Montelione. Structural similarities between SARS-CoV2 3CLpro and other viral proteases suggest potential lead molecules for developing broad spectrum antivirals. Frontiers in Chemistry. 2022; 10: 948553.
Hu K, Lee W, Montelione GT, Sgourakis NG, Vögeli B. Computational approaches for interpreting experimental data and understanding protein structure, dynamics and function relationships. Frontiers in Molecular Biosciences. 2022;9.
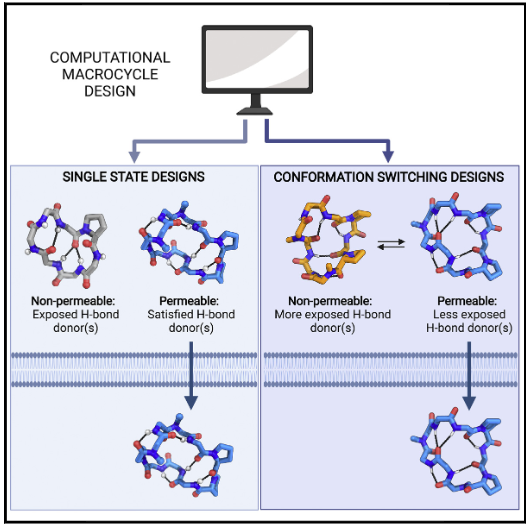
Bhardwaj, Gaurav, O’Connor, Jacob, Rettie, Stephen, Huang, Yen-Hua, Ramelot, Theresa A., Mulligan, Vikram Khipple, Alpkilic, Gizem Gokce, Palmer, Jonathan, Bera, Asim K., Bick, Matthew J., Di Piazza, Maddalena, Li, Xinting, Hosseinzadeh, Parisa, Craven, Timothy W., Tejero, Roberto, Lauko, Anna, Choi, Ryan, Glynn, Calina, Dong, Linlin, Griffin, Robert, van Voorhis, Wesley C., Rodriguez, Jose, Stewart, Lance, Montelione, Gaetano T., Craik, David, and Baker, David. Accurate de novo design of membrane-traversing macrocycles. Cell. 2022 Sep 15;185(19):3520-32.
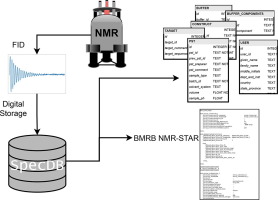
Keith Jeffrey Fraga, Yuanpeng J Huang, Theresa A Ramelot, GVT Swapna, Arwin Lashawn Anak Kendary, Ethan Li, Ian Korf, Gaetano T Montelione. SpecDB: A relational database for archiving biomolecular NMR spectral data. Journal of Magnetic Resonance. 2022. 107268, ISSN 1090-7807.
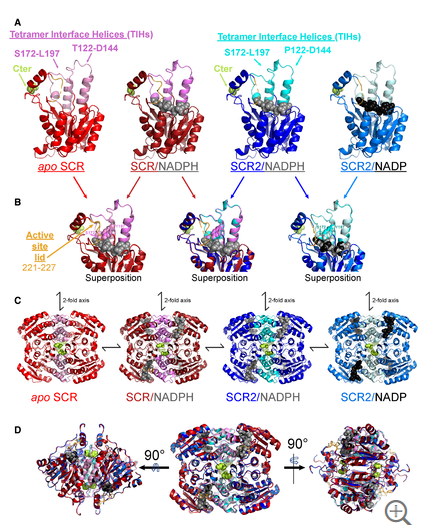
Yaohui Li, Rongzhen Zhang, Chi Wang, Farhad Forouhar, Oliver B Clarke, Sergey Vorobiev, Shikha Singh, Gaetano T Montelione, Thomas Szyperski, Yan Xu, John F Hunt. Oligomeric interactions maintain active-site structure in a noncooperative enzyme family. The EMBO Journal. 2022, 41: e108368.
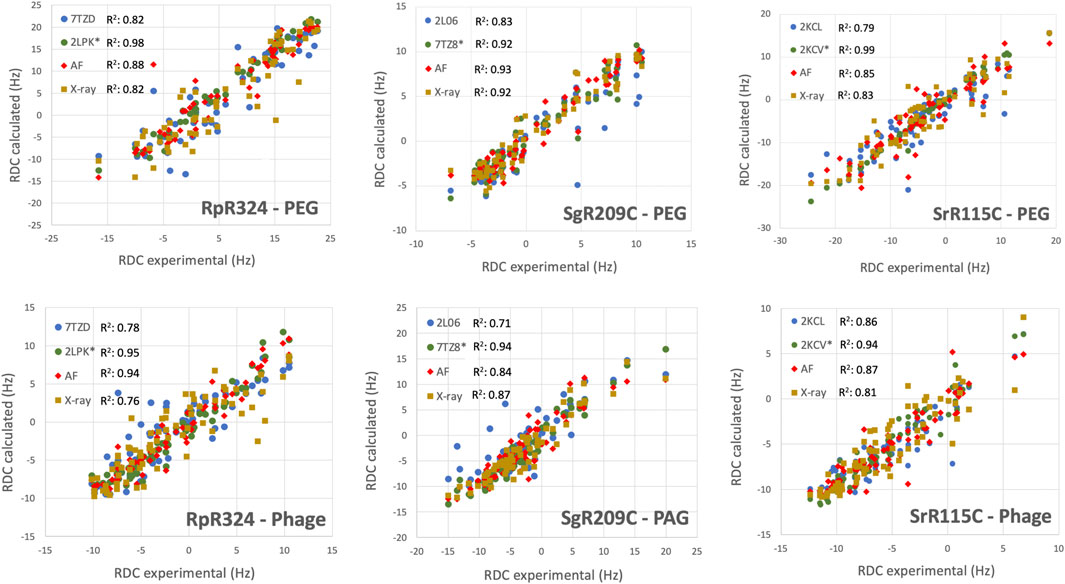
Roberto Tejero, Yuanpeng Janet Huang, Theresa A Ramelot, Gaetano T Montelione. AlphaFold Models of Small Proteins Rival the Accuracy of Solution NMR Structures. Frontiers in Molecular Biosciences. 9, 877000. 2022.
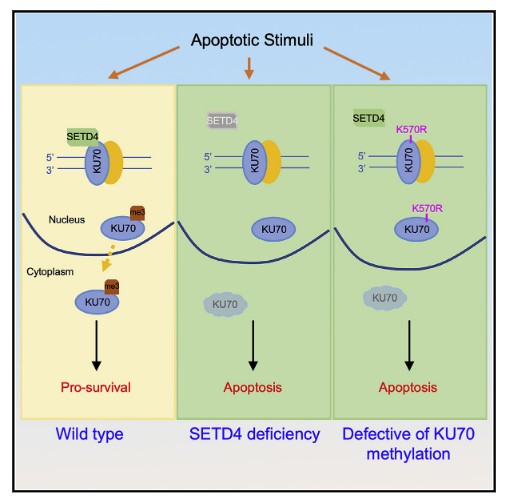
Yuan Wang, Bochao Liu, Huimei Lu, Jingmei Liu, Peter J Romanienko, Gaetano T Montelione, Zhiyuan Shen. SETD4-mediated KU70 methylation suppresses apoptosis. Cell Reports. 2022, 39, 110794.
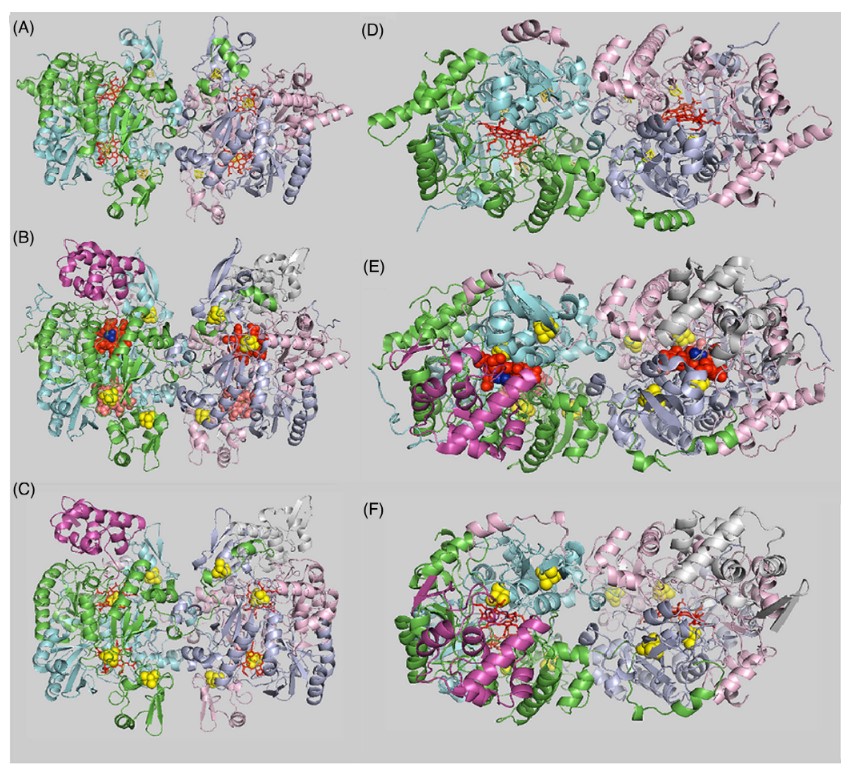
Daniel R Colman, Gilles Labesse, Gurla VT Swapna, Johanna Stefanakis, Gaetano T Montelione, Eric S Boyd, Catherine A Royer. Structural evolution of the ancient enzyme, dissimilatory sulfite reductase. Proteins: Structure, Function, and Bioinformatics. 2022,90:1331–1345.
2021
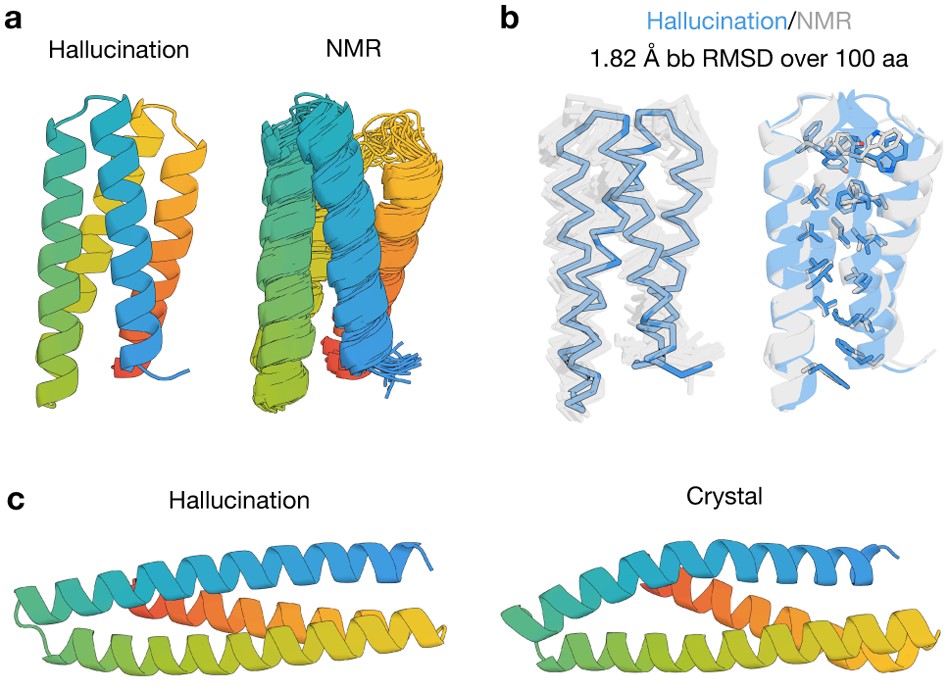
Anishchenko I; Pellock SJ; Chidyausiku TM; Ramelot TA,;Ovchinnikov S; Hao J; Bafna K; Norn C; Kang A; Bera AK; DiMaio F; Carter L; Chow CM; Montelione GT; Baker D. De novo protein design by deep network hallucination. Nature, 2021, 600: 547–552.
Huang YJ, Zhang N, Bersch B, Fidelis K, Inouye M, Ishida Y, Kryshtafovych A, Kobayashi N, Kuroda Y, Liu G, LiWang A, Swapna GVT, Wu N, Yamazaki T, Montelione GT. Assessment of prediction methods for protein structures determined by NMR in CASP14: Impact of AlphaFold2. Proteins: Structure, Function, and Bioinformatics. 2021 Dec;89(12):1959-76..
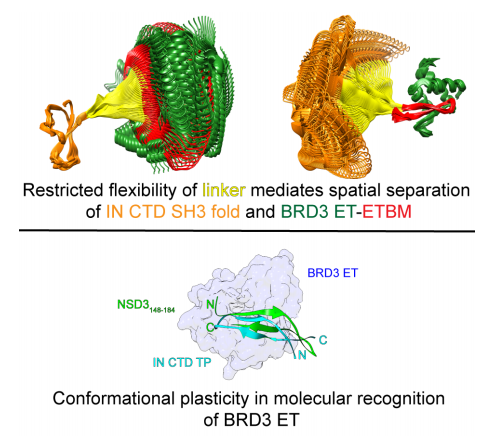
Aiyer S, Swapna GV, Ma LC, Liu G, Hao J, Chalmers G, Jacobs BC, Montelione GT, Roth MJ. A common binding motif in the ET domain of BRD3 forms polymorphic structural interfaces with host and viral proteins. Structure. 2021 Aug 5;29(8):886-98.
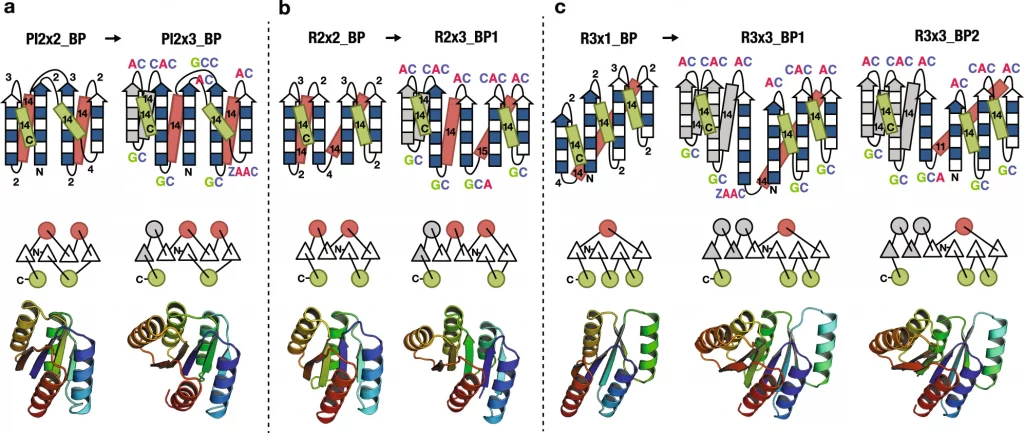
Koga N; Koga R; Liu G; Castellanos J; Montelione GT; Baker D. Role of backbone strain in de novo design of complex alpha/beta protein structures. Nature Communication. 2021, 12: 3921.
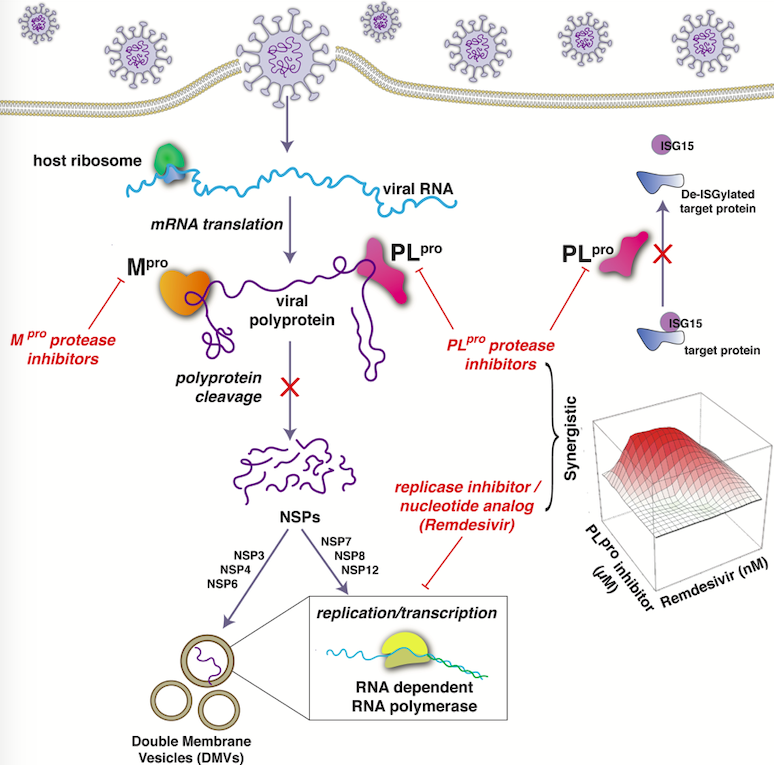
Bafna K; White K; Harish B; Rosales R; Ramelot TA; Acton TB; Moreno E; Kehrer T; Miorin L; Royer CA; Garcia-Sastre A; Krug RM; Montelione GT. Hepatitis C virus drugs that inhibit SARS-CoV-2 papain-like protease synergize with remdesivir to suppress viral replication in cell culture. Cell Reports. 2021, 35: 109133.
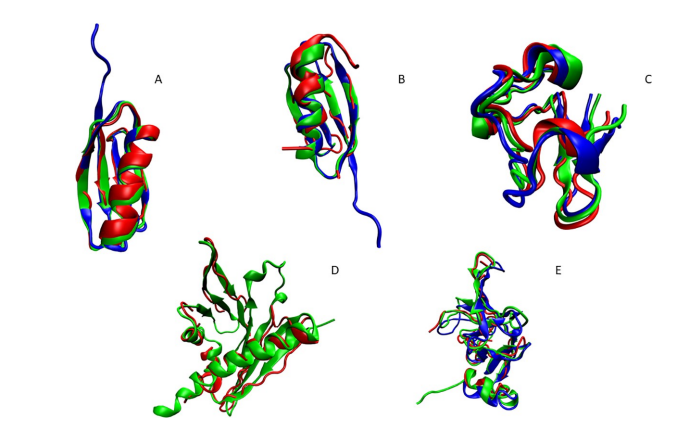
Cole CA, Daigham NS, Liu G, Montelione GT, Valafar H. REDCRAFT: A computational platform using residual dipolar coupling NMR data for determining structures of perdeuterated proteins in solution. PLoS computational biology. 2021 Feb 1;17(2):e1008060.
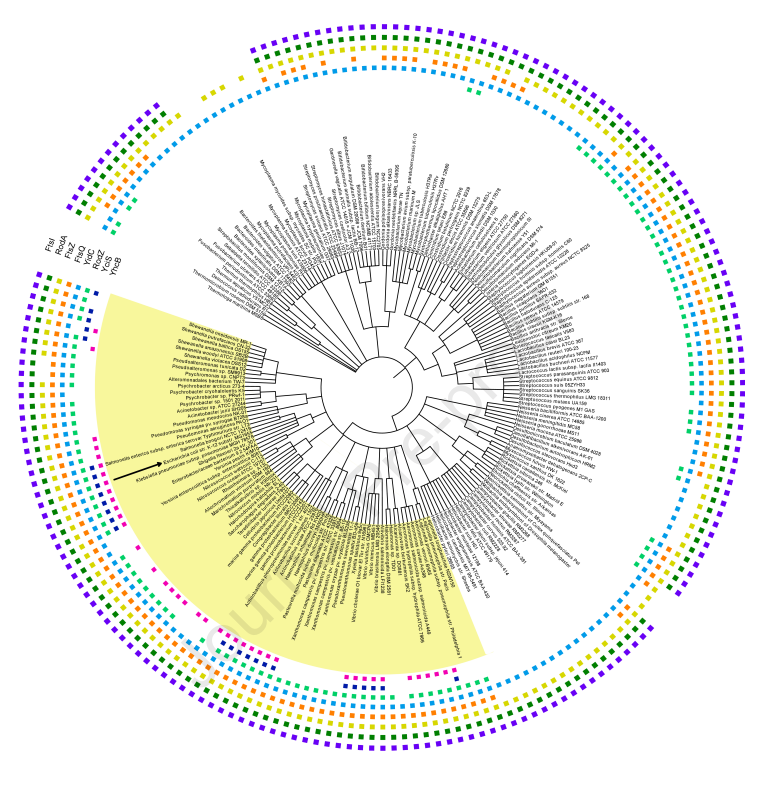
Mehla J, Liechti G, Morgenstein RM, Caufield JH, Hosseinnia A, Gagarinova A, Phanse S, Goodacre N, Brockett M, Sakhawalkar N, Babu M, Xiao Rong, Montelione GT, Vorobiev S, den Blaauwen T, Hunt JF, Uetz P. ZapG (YhcB/DUF1043), a novel cell division protein in gamma-proteobacteria linking the Z-ring to septal peptidoglycan synthesis. Journal of Biological Chemistry. 2021 Jan 1;296.
2020

Maisuradze GG, Montelione GT, Rackovsky S, Skolnick J. Tribute to Harold A. Scheraga. The Journal of Physical Chemistry B. 2020 Oct 23;124(46):10301-2.
Bafna K; Krug RM; Montelione GT. Structural similarity of SARS-CoV2 Mpro and HCV NS3/4A proteases suggests new approaches for identifying existing drugs useful as COVID-19 therapeutics. ChemRxiv. 2020. (Preprint Version)
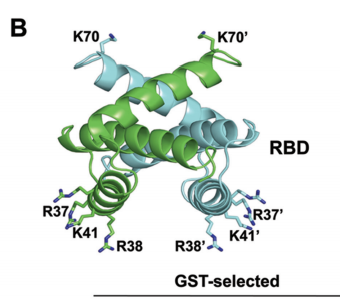
Chen, G., Ma, L.-C., Wang, S., Woltz, R.L., Grasso, E.M., Montelione, G.T., Krug, R.M. A double-stranded RNA platform is required for the interaction between a host restriction factor and the NS1 protein of influenza A virus. Nucleic Acids Research. 48: 304-315, 2020.
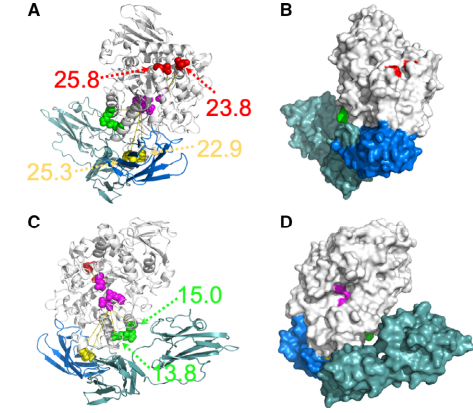
Wang X, Jing X, Deng Y, Nie Y, Xu F, Xu Y, Zhao YL, Hunt JF, Montelione GT, Szyperski T. Evolutionary coupling saturation mutagenesis: Coevolution‐guided identification of distant sites influencing Bacillus naganoensis pullulanase activity. FEBS letters. 2020 Mar;594(5):799-812.